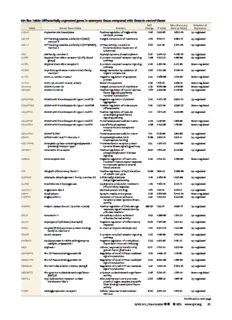
Table Of ContentOn-lineTable:Differentiallyexpressedgenesinaneurysmtissuecomparedwiththoseincontroltissue
Fold FalseDiscovery Directionof
Gene EntrezGeneName Function Change PValue Rate(qValue) Expression
AADAC Arylacetamidedeacetylase Positiveregulationoftriglyceride 4.46 1.33E-05 2.60E-04 Up-regulated
catabolicprocess
ABCA6 ATP-bindingcassette,subfamilyA(ABC1), Integralcomponentofmembrane 3.79 9.15E-14 8.88E-12 Up-regulated
member6
ABCC3 ATP-bindingcassette,subfamilyC(CFTR/MRP), ATPaseactivity,coupledto 6.63 1.21E-10 7.33E-09 Up-regulated
member3 transmembranemovementof
substances
ABI3 ABIfamily,member3 Peptidyl-tyrosinephosphorylation 6.47 2.47E-05 4.56E-04 Up-regulated
ACKR1 Atypicalchemokinereceptor1(Duffyblood G-protein–coupledreceptorsignaling 3.80 7.95E-10 4.18E-08 Up-regulated
group) pathway
ACKR2 Atypicalchemokinereceptor2 G-protein–coupledreceptorsignaling 0.42 3.29E-04 4.41E-03 Down-regulated
pathway
ACSM1 Acyl-CoAsynthetasemedium-chainfamily Energyderivationbyoxidationof 9.87 1.70E-08 6.52E-07 Up-regulated
member1 organiccompounds
ACTC1 Actin,(cid:2),cardiacmuscle1 Negativeregulationofapoptotic 0.30 7.96E-06 1.65E-04 Down-regulated
process
ACTG2 Actin,(cid:3)2,smoothmuscle,enteric Bloodmicroparticle 0.29 1.61E-16 2.36E-14 Down-regulated
ADAM33 ADAMdomain33 Integralcomponentofmembrane 0.23 9.74E-09 3.95E-07 Down-regulated
ADAM8 ADAMdomain8 Positiveregulationoftumornecrosis 4.69 2.93E-04 4.01E-03 Up-regulated
factor(ligand)superfamily
member11production
ADAMTS18 ADAMwiththrombospondintype1motif18 Negativeregulationofplatelet 6.27 4.37E-06 9.85E-05 Up-regulated
aggregation
ADAMTS20 ADAMwiththrombospondintype1motif20 Positiveregulationofmelanocyte 0.18 1.50E-04 2.29E-03 Down-regulated
differentiation
ADAMTS3 ADAMwiththrombospondintype1motif3 Positiveregulationofvascular 3.14 1.67E-06 4.23E-05 Up-regulated
endothelialgrowthfactor
signalingpathway
ADAMTS6 ADAMwiththrombospondintype1motif6 Proteinaceousextracellularmatrix 0.48 1.25E-04 1.96E-03 Down-regulated
ADAMTS8 ADAMwiththrombospondintype1motif8 Low-affinityphosphate 0.40 4.42E-07 1.31E-05 Down-regulated
transmembranetransporter
activity
ADAMTSL1 ADAMTS-like1 Proteinaceousextracellularmatrix 3.13 7.12E-09 3.01E-07 Up-regulated
ADAP2 ArfGAPwithdualPHdomains2 Phosphatidylinositol-3,4,5- 10.88 5.26E-14 5.21E-12 Up-regulated
trisphosphatebinding
ADCYAP1R1 Adenylatecyclase-activatingpolypeptide1 Transmembranereceptorprotein 2.12 4.27E-05 7.45E-04 Up-regulated
(pituitary)receptortypeI tyrosinekinasesignalingpathway
ADORA3 AdenosineA3receptor Positiveregulationof 12.30 1.95E-07 6.14E-06 Up-regulated
phosphatidylinositol3-kinase
signaling
ADRA1A Adrenoceptor(cid:2)1A Negativeregulationofheartrate 0.39 1.79E-05 3.39E-04 Down-regulated
involvedinbaroreceptorresponse
toincreasedsystemicarterial
bloodpressure
AIF1 Allograftinflammatoryfactor1 PositiveregulationofG/Stransition 12.40 1.81E-22 6.08E-20 Up-regulated
1
ofmitoticcellcycle
ALDH1A2 Aldehydedehydrogenase1family,memberA2 3-Chloroallylaldehyde 2.46 2.46E-10 1.42E-08 Up-regulated
dehydrogenaseactivity
ALOX5 Arachidonate5-lipoxygenase Leukotrieneproductioninvolvedin 5.22 7.56E-15 9.12E-13 Up-regulated
inflammatoryresponse
AMOTL2 Angiomotin-like2 Identicalproteinbinding 2.63 1.14E-12 9.56E-11 Up-regulated
AMPH Amphiphysin Synapticvesicleendocytosis 0.45 3.64E-05 6.44E-04 Down-regulated
ANGPT4 Angiopoietin4 Activationoftransmembrane 2.37 5.52E-05 9.32E-04 Up-regulated
receptorproteintyrosinekinase
activity
ANKRD1 Ankyrinrepeatdomain1(cardiacmuscle) PositiveregulationofDNAdamage 206.36 1.13E-18 2.48E-16 Up-regulated
response,signaltransductionby
p53classmediator
ANO3 Anoctamin3 Intracellularcalcium-activated 9.30 4.58E-07 1.35E-05 Up-regulated
chloridechannelactivity
AOAH Acyloxyacylhydrolase(neutrophil) Negativeregulationofinflammatory 12.42 7.97E-07 2.21E-05 Up-regulated
response
APBA2 Amyloid(cid:4)(A4)precursorprotein-binding, Inuteroembryonicdevelopment 3.77 4.55E-06 1.02E-04 Up-regulated
familyA,member2
APLNR Apelinreceptor G-protein–coupledreceptorsignaling 3.53 1.40E-05 2.72E-04 Up-regulated
pathway
APOBEC1 ApolipoproteinBmRNAeditingenzyme, Negativeregulationofmethylation- 8.02 4.81E-07 1.41E-05 Up-regulated
catalyticpolypeptide1 dependentchromatinsilencing
ARG1 Arginase1 Cellularresponsetotransforming 12.10 1.70E-05 3.25E-04 Up-regulated
growthfactor(cid:4)stimulus
ARHGAP18 RhoGTPase-activatingprotein18 RegulationofsmallGTPase-mediated 2.79 5.98E-07 1.71E-05 Up-regulated
signaltransduction
ARHGAP30 RhoGTPase-activatingprotein30 RegulationofsmallGTPase-mediated 6.44 8.32E-06 1.70E-04 Up-regulated
signaltransduction
ARHGDIB RhoGDP-dissociationinhibitor(GDI)(cid:4) RegulationofsmallGTPase-mediated 2.27 5.35E-04 6.67E-03 Up-regulated
signaltransduction
ARHGEF26 Rhoguaninenucleotideexchangefactor (cid:5)-Guanyl-nucleotideexchangefactor 0.46 6.31E-06 1.37E-04 Down-regulated
(GEF)26 activity
ARNTL2 Arylhydrocarbonreceptornuclear RNApolymeraseIIcorepromoter 15.63 3.60E-21 1.09E-18 Up-regulated
translocator-like2 proximalregionsequence-specific
DNA-bindingtranscriptionfactor
activity
ASGR1 Asialoglycoproteinreceptor1 Cellularresponsetoextracellular 16.83 3.43E-24 1.54E-21 Up-regulated
stimulus
Continuedonnextpage
AJNRAmJNeuroradiol●:● ●2015 www.ajnr.org E1
On-lineTable:Continued
Fold FalseDiscovery Directionof
Gene EntrezGeneName Function Change PValue Rate(qValue) Expression
ATF3 Activatingtranscriptionfactor3 RNApolymeraseIItranscription 0.43 1.06E-06 2.85E-05 Down-regulated
regulatoryregionsequence-
specificDNA-binding
transcriptionfactoractivity
involvedinpositiveregulationof
transcription
ATP10B ATPase,classV,type10B Phospholipid-translocatingATPase 0.12 5.81E-05 9.67E-04 Down-regulated
activity
ATP1A2 ATPase,Na(cid:2)/K(cid:2)transporting,(cid:2)2polypeptide Regulationofrespiratorygaseous 0.46 3.44E-07 1.04E-05 Down-regulated
exchangebyneurologicsystem
process
ATP2A3 ATPase,Ca(cid:2)(cid:2)transporting,ubiquitous Platelet-densetubularnetwork 0.37 1.07E-12 9.10E-11 Down-regulated
membrane
ATP6V0D2 ATPase,H(cid:2)transporting,lysosomal38kDa, Vacuolarproton-transportingV-type 6.09 4.10E-05 7.16E-04 Up-regulated
V0subunitd2 ATPasecomplex
ATP8B4 ATPase,classI,type8B,member4 Phospholipid-translocatingATPase 6.21 8.22E-06 1.69E-04 Up-regulated
activity
BAG3 BCL2-associatedathanogene3 Extrinsicapoptoticsignalingpathway 0.50 1.33E-06 3.49E-05 Down-regulated
viadeath-domainreceptors
BASP1 Brain-abundant,membrane-attachedsignal Positiveregulationofmetanephric 4.18 6.71E-08 2.36E-06 Up-regulated
protein1 uretericbuddevelopment
BATF Basicleucinezippertranscriptionfactor, DNAdamageresponse,signal 11.20 1.40E-04 2.15E-03 Up-regulated
ATF-like transductionbyp53class
mediator
BCHE Butyrylcholinesterase Negativeregulationofsynaptic 0.23 2.09E-12 1.73E-10 Down-regulated
transmission
BHLHE22 Basichelix-loop-helixfamily,membere22 Sequence-specificDNAbindingRNA 3.21 6.64E-04 7.97E-03 Up-regulated
polymeraseIItranscriptionfactor
activity
BIN2 Bridgingintegrator2 Phagocytosis,engulfment 6.63 4.30E-05 7.47E-04 Up-regulated
BIRC3 BaculoviralIAPrepeatcontaining3 Regulationofnucleotide-binding 2.18 7.96E-04 9.39E-03 Up-regulated
oligomerizationdomain
containingsignalingpathway
BLNK B-celllinker Transmembranereceptorprotein 14.87 3.80E-08 1.38E-06 Up-regulated
tyrosinekinasesignalingpathway
BMPR1B Bonemorphogeneticproteinreceptor,typeIB Positiveregulationofextrinsic 3.03 7.27E-08 2.54E-06 Up-regulated
apoptoticsignalingpathwayvia
death-domainreceptors
BMX BMXnonreceptortyrosinekinase Cellularcomponentdisassembly 0.46 2.41E-04 3.43E-03 Down-regulated
involvedinexecutionphaseof
apoptosis
BNIPL BCL2/adenovirusE1B19kDainteracting Negativeregulationofcell 9.55 1.92E-06 4.75E-05 Up-regulated
protein-like proliferation
BOC BOCcelladhesionassociated,oncogene Positiveregulationofmusclecell 2.46 2.82E-10 1.61E-08 Up-regulated
regulated differentiation
BTK Brutonagammaglobulinemiatyrosinekinase PositiveregulationofNF-(cid:6)B 7.30 9.06E-07 2.47E-05 Up-regulated
transcriptionfactoractivity
BVES Bloodvesselepicardialsubstance Substrateadhesion-dependentcell 0.34 1.20E-07 4.02E-06 Down-regulated
spreading
C11orf16 Chromosome11openreadingframe16 — 7.33 2.48E-04 3.51E-03 Up-regulated
C1orf110 Chromosome1openreadingframe110 — 2.35 1.16E-04 1.84E-03 Up-regulated
C1orf162 Chromosome1openreadingframe162 Integralcomponentofmembrane 60.67 2.05E-12 1.71E-10 Up-regulated
C1orf54 Chromosome1openreadingframe54 Extracellularregion 3.66 5.77E-08 2.04E-06 Up-regulated
C1QA Complementcomponent1,qsubcomponent, Complementactivation,classic 4.20 1.24E-15 1.64E-13 Up-regulated
Achain pathway
C1QB Complementcomponent1,qsubcomponent, Complementactivation,classic 5.21 1.19E-11 8.50E-10 Up-regulated
Bchain pathway
C1QC Complementcomponent1,qsubcomponent, Negativeregulationofgranulocyte 5.26 3.69E-14 3.87E-12 Up-regulated
Cchain differentiation
C1QTNF3 C1qandtumornecrosisfactor–related Negativeregulationofmonocyte 0.49 2.70E-06 6.41E-05 Down-regulated
protein3 chemotacticprotein1production
C1QTNF9 C1qandtumornecrosisfactor–related Extracellularregion 2.65 8.54E-04 9.97E-03 Up-regulated
protein9
C1RL Complementcomponent1,rsubcomponent- Complementactivation,classic 2.01 3.41E-06 7.88E-05 Up-regulated
like pathway
C1S Complementcomponent1,ssubcomponent Complementactivation,classic 2.01 3.39E-06 7.84E-05 Up-regulated
pathway
C2orf40 Chromosome2openreadingframe40 Cyclincatabolicprocess 4.57 5.21E-09 2.29E-07 Up-regulated
C3AR1 Complementcomponent3areceptor1 Positiveregulationvascular 7.28 3.84E-09 1.74E-07 Up-regulated
endothelialgrowthfactor
production
C8orf34 Chromosome8openreadingframe34 — 13.83 1.13E-09 5.81E-08 Up-regulated
CA3 CarbonicanhydraseIII,musclespecific Smallmoleculemetabolicprocess 0.20 5.66E-05 9.52E-04 Down-regulated
CA9 CarbonicanhydraseIX RegulationoftranscriptionfromRNA 0.15 1.63E-16 2.36E-14 Down-regulated
polymeraseIIpromoterin
responsetohypoxia
CABLES1 Cdk5andAblenzymesubstrate1 Cyclin-dependentproteinserine/ 2.23 4.16E-04 5.38E-03 Up-regulated
threoninekinaseregulatoractivity
CADM1 Celladhesionmolecule1 Positiveregulationofnaturalkiller 2.26 2.76E-04 3.84E-03 Up-regulated
cell–mediatedcytotoxicity
CADM3 Celladhesionmolecule3 Proteinhomodimerizationactivity 0.37 1.92E-09 9.28E-08 Down-regulated
CAPG Cappingprotein(actinfilament),gelsolin-like Barbed-endactinfilamentcapping 2.17 9.52E-06 1.92E-04 Up-regulated
Continuedonnextpage
E2 Holcomb ●2015 www.ajnr.org
On-lineTable:Continued
Fold FalseDiscovery Directionof
Gene EntrezGeneName Function Change PValue Rate(qValue) Expression
CASP1 Caspase1,apoptosis-relatedcysteine Nucleotide-bindingdomain,leucine- 11.56 1.02E-05 2.05E-04 Up-regulated
peptidase richrepeatcontainingreceptor
signalingpathway
CASP8 Caspase8,apoptosis-relatedcysteine Positiveregulationofprotein 4.88 7.45E-05 1.22E-03 Up-regulated
peptidase insertionintomitochondrial
membraneinvolvedinapoptotic
signalingpathway
CASQ2 Calsequestrin2(cardiacmuscle) Regulationofcardiacmuscle 0.21 3.21E-15 3.93E-13 Down-regulated
contractionbyregulationofthe
releaseofsequesteredcalcium
ion
CCBE1 CollagenandcalciumbindingEGFdomains1 Positiveregulationofvascular 4.05 6.12E-07 1.74E-05 Up-regulated
endothelialgrowthfactor
signalingpathway
CCDC102B Coiled-coildomaincontaining102B — 3.75 2.20E-06 5.33E-05 Up-regulated
CCDC126 Coiled-coildomaincontaining126 (cid:2)-1,6-Mannosylglycoprotein6-(cid:4)-N- 2.03 1.30E-04 2.03E-03 Up-regulated
acetylglucosaminyltransferase
activity
CCL13 Chemokine(C-Cmotif)ligand13 Cellularcalciumionhomeostasis 8.67 6.98E-06 1.48E-04 Up-regulated
CCL14 Chemokine(C-Cmotif)ligand14 Positiveregulationofcell 2.43 1.30E-06 3.45E-05 Up-regulated
proliferation
CCL19 Chemokine(C-Cmotif)ligand19 Positiveregulationofdendriticcell 7.04 1.37E-05 2.67E-04 Up-regulated
antigenprocessingand
presentation
CCL2 Chemokine(C-Cmotif)ligand2 G-protein–coupledreceptorsignaling 5.38 1.58E-10 9.30E-09 Up-regulated
pathway,coupledtocyclic
nucleotidesecondmessenger
CCL21 Chemokine(C-Cmotif)ligand21 Positiveregulationofdendriticcell 8.19 2.35E-50 7.39E-47 Up-regulated
antigenprocessingand
presentation
CCR1 Chemokine(C-Cmotif)receptor1 G-protein–coupledreceptorsignaling 32.11 8.32E-14 8.16E-12 Up-regulated
pathway,coupledtocyclic
nucleotidesecondmessenger
CCR2 Chemokine(C-Cmotif)receptor2 Positiveregulationofimmune 13.14 8.62E-07 2.36E-05 Up-regulated
complexclearancebymonocytes
andmacrophages
CCR5 Chemokine(C-Cmotif)receptor5 Releaseofsequesteredcalciumion 24.10 9.71E-12 7.09E-10 Up-regulated
(gene/pseudogene) intocytosolbysarcoplasmic
reticulum
CD14 CD14molecule Positiveregulationoftumornecrosis 9.22 1.39E-17 2.33E-15 Up-regulated
factorproduction
CD163 CD163molecule Integralcomponentofplasma 2.73 2.43E-07 7.48E-06 Up-regulated
membrane
CD180 CD180molecule Positiveregulationof 13.45 2.66E-11 1.80E-09 Up-regulated
lipopolysaccharide-mediated
signalingpathway
CD1C CD1cmolecule T-cellactivationinvolvedinimmune 7.40 3.69E-04 4.90E-03 Up-regulated
response
CD200 CD200molecule Integralcomponentofplasma 2.30 2.80E-09 1.31E-07 Up-regulated
membrane
CD300LG CD300molecule-likefamilymemberg Extracellularvesicularexosome 2.82 3.48E-05 6.21E-04 Up-regulated
CD34 CD34molecule Positiveregulationofglialcellline– 0.44 2.10E-08 7.96E-07 Down-regulated
derivedneurotrophicfactor
secretion
CD48 CD48molecule Anchoredcomponentofplasma 8.55 3.47E-08 1.27E-06 Up-regulated
membrane
CD53 CD53molecule Positiveregulationofmyoblast 5.49 1.65E-06 4.20E-05 Up-regulated
fusion
CD68 CD68molecule Cellularresponsetoorganic 4.46 8.29E-09 3.43E-07 Up-regulated
substance
CD74 CD74molecule,MHCclassIIinvariantchain Negativeregulationofintrinsic 39.56 7.67E-18 1.36E-15 Up-regulated
apoptoticsignalingpathwayin
responsetoDNAdamagebyp53
classmediator
CD80 CD80molecule Positiveregulationofgranulocyte- 15.17 1.39E-16 2.07E-14 Up-regulated
macrophagecolony-stimulating
factorbiosyntheticprocess
CD83 CD83molecule PositiveregulationofCD4-positive, 13.66 4.69E-09 2.08E-07 Up-regulated
(cid:2)-(cid:4)T-celldifferentiation
CD84 CD84molecule Integralcomponentofplasma 11.44 1.40E-14 1.59E-12 Up-regulated
membrane
CD86 CD86molecule Positiveregulationofinterleukin-2 12.86 8.56E-11 5.24E-09 Up-regulated
biosyntheticprocess
CD93 CD93molecule Cytoplasmicmembrane-bounded 2.67 2.17E-10 1.26E-08 Up-regulated
vesicle
CDH11 Cadherin11,type2,OB-cadherin(osteoblast) Corticospinaltractmorphogenesis 4.57 1.32E-27 8.90E-25 Up-regulated
CDH19 Cadherin19,type2 Integralcomponentofmembrane 0.36 1.73E-04 2.59E-03 Down-regulated
CDH2 Cadherin2,type1,N-cadherin(neuronal) NegativeregulationofcanonicalWnt 6.97 4.46E-12 3.50E-10 Up-regulated
signalingpathway
CDH20 Cadherin20,type2 Integralcomponentofmembrane 3.71 4.35E-05 7.51E-04 Up-regulated
CDH3 Cadherin3,type1,P-cadherin(placental) Positiveregulationofinsulin-like 9.70 8.01E-05 1.30E-03 Up-regulated
growthfactorreceptorsignaling
pathway
CDH6 Cadherin6,type2,K-cadherin(fetalkidney) Cell–celljunctionorganization 3.83 3.09E-06 7.21E-05 Up-regulated
Continuedonnextpage
AJNRAmJNeuroradiol●:● ●2015 www.ajnr.org E3
On-lineTable:Continued
Fold FalseDiscovery Directionof
Gene EntrezGeneName Function Change PValue Rate(qValue) Expression
CDH8 Cadherin8,type2 Synaptictransmission,glutamatergic 0.34 2.55E-04 3.59E-03 Down-regulated
CDK15 Cyclin-dependentkinase15 Cyclin-dependentproteinserine/ 0.36 6.08E-04 7.47E-03 Down-regulated
threoninekinaseactivity
CDKN1A Cyclin-dependentkinaseinhibitor1A(p21,Cip1) DNA-damageresponse,signal 3.78 3.89E-14 4.02E-12 Up-regulated
transductionbyp53class
mediatorresultingincell-cycle
arrest
CDKN2A Cyclin-dependentkinaseinhibitor2A PositiveregulationofDNA-damage 54.19 1.81E-20 4.88E-18 Up-regulated
response,signaltransductionby
p53classmediator
CDKN2B Cyclin-dependentkinaseinhibitor2B Positiveregulationoftransforming 4.87 5.35E-10 2.91E-08 Up-regulated
(p15,inhibitsCDK4) growthfactor(cid:4)receptorsignaling
pathway
CELSR2 Cadherin,EGFLAGseven-passG-type Neuralplateanterior/posterior 0.49 2.92E-04 4.01E-03 Down-regulated
receptor2 regionalization
CFH ComplementfactorH Complementactivation,alternative 6.34 2.41E-26 1.33E-23 Up-regulated
pathway
CFP Complementfactorproperdin Complementactivation,alternative 5.33 4.32E-06 9.76E-05 Up-regulated
pathway
CH25H Cholesterol25-hydroxylase Cholesterol25-hydroxylaseactivity 4.67 7.58E-11 4.73E-09 Up-regulated
CHAC1 ChaC,cationtransportregulatorhomolog1 Intrinsicapoptoticsignalingpathway 4.45 3.16E-05 5.70E-04 Up-regulated
(Escherichiacoli) inresponsetoendoplasmic
reticulumstress
CHI3L1 Chitinase3-like1(cartilageglycoprotein39) Positiveregulationofpeptidyl- 7.97 5.85E-13 5.10E-11 Up-regulated
threoninephosphorylation
CHI3L2 Chitinase3-like2 Carbohydratemetabolicprocess 55.72 1.47E-07 4.77E-06 Up-regulated
CHIT1 Chitinase1(chitotriosidase) Polysaccharidecatabolicprocess 24.30 2.13E-11 1.47E-09 Up-regulated
CHMP4C Chargedmultivesicularbodyprotein4C Positiveregulationofviralrelease 0.34 1.35E-04 2.09E-03 Down-regulated
fromhostcell
CHN2 Chimerin2 RegulationofsmallGTPase-mediated 3.47 7.42E-05 1.22E-03 Up-regulated
signaltransduction
CHP2 Calcineurin-likeEF-handprotein2 Positiveregulationoftranscription 2.64 2.30E-05 4.29E-04 Up-regulated
fromRNApolymeraseIIpromoter
CHRDL2 Chordin-like2 Cartilagedevelopment 6.46 1.29E-05 2.54E-04 Up-regulated
CLEC12A C-typelectindomainfamily12,memberA Integralcomponentofmembrane 20.14 7.62E-10 4.05E-08 Up-regulated
CLEC4A C-typelectindomainfamily4,memberA Transmembranesignalingreceptor 8.82 1.34E-07 4.39E-06 Up-regulated
activity
CLEC4D C-typelectindomainfamily4,memberD Integralcomponentofmembrane 8.99 5.31E-08 1.89E-06 Up-regulated
CLEC4E C-typelectindomainfamily4,memberE Positiveregulationofcytokine 21.53 5.81E-13 5.10E-11 Up-regulated
secretion
CLEC7A C-typelectindomainfamily7,memberA Cellsurfacepatternrecognition 13.53 1.16E-17 2.00E-15 Up-regulated
receptorsignalingpathway
CMTM1 CKLF-likeMARVELtransmembranedomain Integralcomponentofmembrane 3.59 5.48E-04 6.82E-03 Up-regulated
containing1
COL10A1 Collagen,typeX,(cid:2)1 Proteinaceousextracellularmatrix 163.48 3.66E-26 1.91E-23 Up-regulated
COL11A1 Collagen,typeXI,(cid:2)1 Detectionofmechanicalstimulus 29.94 7.85E-45 1.48E-41 Up-regulated
involvedinsensoryperceptionof
sound
COL12A1 Collagen,typeXII,(cid:2)1 Extracellularmatrixstructural 3.28 1.11E-16 1.68E-14 Up-regulated
constituentconferringtensile
strength
COL1A1 Collagen,typeI,(cid:2)1 Cartilagedevelopmentinvolvedin 2.09 1.04E-04 1.66E-03 Up-regulated
endochondralbone
morphogenesis
COL1A2 Collagen,typeI,(cid:2)2 Transforminggrowthfactor(cid:4) 2.05 4.84E-05 8.26E-04 Up-regulated
receptorsignalingpathway
COL23A1 Collagen,typeXXIII,(cid:2)1 Extracellularmatrixorganization 8.57 1.65E-23 6.49E-21 Up-regulated
COL25A1 Collagen,typeXXV,(cid:2)1 Integralcomponentofplasma 2.30 7.73E-04 9.15E-03 Up-regulated
membrane
COL28A1 Collagen,typeXXVIII,(cid:2)1 Negativeregulationof 0.29 6.27E-06 1.36E-04 Down-regulated
endopeptidaseactivity
COL4A3 Collagen,typeIV,(cid:2)3(Goodpastureantigen) Activationofcysteine-type 3.97 1.21E-11 8.56E-10 Up-regulated
endopeptidaseactivityinvolvedin
apoptoticprocess
COL4A4 Collagen,typeIV,(cid:2)4 Extracellularmatrixstructural 5.86 8.57E-15 1.01E-12 Up-regulated
constituent
COL8A1 Collagen,typeVIII,(cid:2)1 Positiveregulationofcell-substrate 2.26 1.36E-07 4.45E-06 Up-regulated
adhesion
COLQ Collagen-liketailsubunit(singlestrandof Acetylcholinecatabolicprocessin 3.56 1.09E-04 1.74E-03 Up-regulated
homotrimer)ofasymmetric synapticcleft
acetylcholinesterase
CORO1A Coronin,actin-bindingprotein1A Homeostasisofnumberofcells 10.29 4.18E-06 9.53E-05 Up-regulated
withinatissue
CP Ceruloplasmin(ferroxidase) Extracellularvesicularexosome 4.48 1.54E-24 7.24E-22 Up-regulated
CREB5 cAMP-responsiveelement-bindingprotein5 RNApolymeraseIIcorepromoter 0.49 2.71E-04 3.78E-03 Down-regulated
proximalregionsequence-specific
DNA-bindingtranscriptionfactor
activityinvolvedinpositive
regulationoftranscription
CRTAC1 Cartilageacidicprotein1 Proteinaceousextracellularmatrix 2.20 4.42E-05 7.60E-04 Up-regulated
CSF1R Colony-stimulatingfactor1receptor Cellularresponsetomacrophage 5.68 9.51E-30 7.47E-27 Up-regulated
colony-stimulatingfactorstimulus
Continuedonnextpage
E4 Holcomb ●2015 www.ajnr.org
On-lineTable:Continued
Fold FalseDiscovery Directionof
Gene EntrezGeneName Function Change PValue Rate(qValue) Expression
CSF2RB Colony-stimulatingfactor2receptor,(cid:4),low Granulocyte-macrophagecolony- 4.70 3.84E-07 1.15E-05 Up-regulated
affinity(granulocyte-macrophage) stimulatingfactorreceptor
complex
CTBS Chitobiase,di-N-acetyl- Oligosaccharidecatabolicprocess 2.10 2.31E-05 4.30E-04 Up-regulated
CTGF Connectivetissuegrowthfactor Positiveregulationofcysteine-type 4.44 2.47E-19 5.96E-17 Up-regulated
endopeptidaseactivityinvolvedin
apoptoticprocess
CTSB CathepsinB Proteolysisinvolvedincellular 2.43 1.39E-08 5.40E-07 Up-regulated
proteincatabolicprocess
CTSE CathepsinE Antigenprocessingandpresentation 3.77 6.26E-04 7.65E-03 Up-regulated
ofexogenouspeptideantigenvia
MHCclassII
CTSK CathepsinK Proteolysisinvolvedincellular 2.04 1.33E-06 3.49E-05 Up-regulated
proteincatabolicprocess
CTSS CathepsinS Antigenprocessingandpresentation 5.95 4.79E-21 1.41E-18 Up-regulated
ofexogenouspeptideantigenvia
MHCclassI,TAPindependent
CX3CR1 Chemokine(C-X3-Cmotif)receptor1 Negativeregulationofextrinsic 61.52 1.33E-23 5.45E-21 Up-regulated
apoptoticsignalingpathwayin
absenceofligand
CXCL16 Chemokine(C-X-Cmotif)ligand16 Low-densitylipoproteinreceptor 2.31 2.63E-06 6.27E-05 Up-regulated
activity
CXCL6 Chemokine(C-X-Cmotif)ligand6 Defenseresponsetobacterium 5.29 5.34E-06 1.18E-04 Up-regulated
CXCL8 Chemokine(C-X-Cmotif)ligand8 Regulationofsingle-strandedviral 12.37 4.62E-07 1.36E-05 Up-regulated
RNAreplicationviadouble-
strandedDNAintermediate
CXCR4 Chemokine(C-X-Cmotif)receptor4 Positiveregulationofcytosolic 7.52 2.09E-04 3.03E-03 Up-regulated
calciumionconcentration
CXorf21 ChromosomeXopenreadingframe21 — 9.42 1.23E-05 2.44E-04 Up-regulated
CXorf36 ChromosomeXopenreadingframe36 Extracellularregion 3.63 9.52E-06 1.92E-04 Up-regulated
CYBB Cytochromeb-245,(cid:4)polypeptide Antigenprocessingandpresentation 3.99 2.08E-10 1.21E-08 Up-regulated
ofexogenouspeptideantigenvia
MHCclassI,TAPdependent
CYP26A1 CytochromeP450,family26,subfamilyA, Negativeregulationofretinoicacid 86.52 3.30E-14 3.50E-12 Up-regulated
polypeptide1 receptorsignalingpathway
CYR61 Cysteine-rich,angiogenicinducer61 Positiveregulationofcysteine-type 3.18 3.15E-15 3.90E-13 Up-regulated
endopeptidaseactivityinvolvedin
apoptoticprocess
CYSLTR1 Cysteinylleukotrienereceptor1 Positiveregulationofcytosolic 4.17 7.78E-04 9.19E-03 Up-regulated
calciumionconcentration
DAAM2 Dishevelled-associatedactivatorof Actincytoskeletonorganization 2.47 1.38E-10 8.29E-09 Up-regulated
morphogenesis2
DAB2 Dab,mitogen-responsivephosphoprotein, Positiveregulationoftransforming 2.44 3.78E-11 2.51E-09 Up-regulated
homolog2(Drosophila) growthfactor(cid:4)receptorsignaling
pathway
DAPK2 Death-associatedproteinkinase2 Regulationofintrinsicapoptotic 3.41 3.45E-08 1.26E-06 Up-regulated
signalingpathway
DAPL1 Death-associatedprotein-like1 Celldifferentiation 2.25 6.95E-04 8.32E-03 Up-regulated
DAPP1 Dualadaptorofphosphotyrosineand3- Phosphatidylinositol-3,4,5- 9.11 7.08E-06 1.49E-04 Up-regulated
phosphoinositides trisphosphatebinding
DCBLD2 Discoidin,CUBandLCCLdomaincontaining2 Intracellularreceptorsignaling 0.44 1.24E-07 4.12E-06 Down-regulated
pathway
DCHS2 Dachsouscadherin-related2 Integralcomponentofmembrane 0.35 2.79E-04 3.86E-03 Down-regulated
DDAH1 Dimethylargininedimethylaminohydrolase1 Positiveregulationofnitric-oxide 2.24 7.24E-05 1.19E-03 Up-regulated
biosyntheticprocess
DES Desmin Structuralconstituentof 0.35 2.39E-12 1.96E-10 Down-regulated
cytoskeleton
DHRS7C Dehydrogenase/reductase(SDRfamily) Regulationofreleaseofsequestered 8.28 1.43E-06 3.73E-05 Up-regulated
member7C calciumionintocytosolby
sarcoplasmicreticulum
DKK3 DickkopfWntsignalingpathwayinhibitor3 Negativeregulationofaldosterone 3.18 7.14E-12 5.42E-10 Up-regulated
biosyntheticprocess
DLG2 Discs,largehomolog2(Drosophila) Negativeregulationofphosphatase 0.14 3.48E-12 2.76E-10 Down-regulated
activity
DLX1 Distal-lesshomeobox1 RegulationoftranscriptionfromRNA 3.28 3.75E-05 6.61E-04 Up-regulated
polymeraseIIpromoterinvolved
inforebrainneuronfate
commitment
DLX5 Distal-lesshomeobox5 RNApolymeraseIIcorepromoter 19.90 3.84E-36 4.52E-33 Up-regulated
proximalregionsequence-specific
DNA-bindingtranscriptionfactor
activityinvolvedinpositive
regulationoftranscription
DLX6 Distal-lesshomeobox6 Transcriptionregulatoryregion 30.78 1.10E-19 2.79E-17 Up-regulated
sequence-specificDNAbinding
DNAH11 Dynein,axonemal,heavy-chain11 Ciliumorflagellum-dependentcell 0.09 1.52E-18 3.25E-16 Down-regulated
motility
DNAJB5 DnaJ(Hsp40)homolog,subfamilyB,member5 Negativeregulationoftranscription 0.50 2.12E-07 6.65E-06 Down-regulated
fromRNApolymeraseIIpromoter
DNAJC6 DnaJ(Hsp40)homolog,subfamilyC,member6 Post-Golgivesicle-mediated 2.13 2.33E-04 3.32E-03 Up-regulated
transport
DNER Delta/notch-likeEGFrepeatcontaining Transmembranesignalingreceptor 0.27 6.92E-09 2.94E-07 Down-regulated
activity
Continuedonnextpage
AJNRAmJNeuroradiol●:● ●2015 www.ajnr.org E5
On-lineTable:Continued
Fold FalseDiscovery Directionof
Gene EntrezGeneName Function Change PValue Rate(qValue) Expression
DOCK2 Dedicatorofcytokinesis2 Myeloiddendriticcellactivation 5.33 1.16E-04 1.84E-03 Up-regulated
involvedinimmuneresponse
DOK3 Dockingprotein3 Rasproteinsignaltransduction 14.55 5.03E-10 2.76E-08 Up-regulated
DPYSL3 Dihydropyrimidinase-like3 Hydrolaseactivity,actingon 2.14 3.65E-09 1.67E-07 Up-regulated
carbonOnitrogen(butnot
peptide)bonds,incyclicamides
DUSP6 Dual-specificityphosphatase6 Regulationoffibroblastgrowth 2.61 1.27E-07 4.21E-06 Up-regulated
factorreceptorsignalingpathway
DYNC1I1 Dynein,cytoplasmic1,intermediate-chain1 Antigenprocessingandpresentation 3.31 2.60E-06 6.21E-05 Up-regulated
ofexogenouspeptideantigenvia
MHCclassII
EGR1 Earlygrowthresponse1 RNApolymeraseIIcorepromoter 2.85 4.04E-11 2.66E-09 Up-regulated
proximalregionsequence-specific
DNA-bindingtranscriptionfactor
activityinvolvedinpositive
regulationoftranscription
EMCN Endomucin Integralcomponentofmembrane 4.16 1.44E-14 1.62E-12 Up-regulated
EMP1 Epithelialmembraneprotein1 Multicellularorganismaldevelopment 2.36 1.01E-08 4.06E-07 Up-regulated
ENPEP Glutamylaminopeptidase(aminopeptidaseA) Regulationofsystemicarterialblood 5.79 2.40E-18 4.75E-16 Up-regulated
pressurebyrenin–angiotensin
ENPP2 Ectonucleotidepyrophosphatase/ Alkylglycerophosphoethanolamine 4.18 1.29E-21 4.06E-19 Up-regulated
phosphodiesterase2 phosphodiesteraseactivity
ENPP3 Ectonucleotidepyrophosphatase/ Phosphate-containingcompound 3.97 3.35E-06 7.76E-05 Up-regulated
phosphodiesterase3 metabolicprocess
EPHA5 EphreceptorA5 Regulationofinsulinsecretion 0.36 1.20E-05 2.39E-04 Down-regulated
involvedincellularresponseto
glucosestimulus
EPSTI1 Epithelialstromalinteraction1(breast) — 3.79 6.98E-04 8.34E-03 Up-regulated
ESM1 Endothelialcell-specificmolecule1 Positiveregulationofhepatocyte 0.45 2.16E-05 4.06E-04 Down-regulated
growthfactorreceptorsignaling
pathway
ETV5 ETSvariant5 RNApolymeraseIItranscription 2.05 1.78E-05 3.37E-04 Up-regulated
regulatoryregionsequence-
specificDNA-binding
transcriptionfactoractivity
involvedinpositiveregulationof
transcription
EVI2A Ecotropicviralintegrationsite2A Transmembranesignalingreceptor 9.44 6.68E-12 5.11E-10 Up-regulated
activity
EVI2B Ecotropicviralintegrationsite2B Integralcomponentofplasma 14.52 5.18E-06 1.15E-04 Up-regulated
membrane
EXOC3L1 Exocystcomplexcomponent3-like1 Peptidehormonesecretion 5.87 1.50E-06 3.87E-05 Up-regulated
F5 CoagulationfactorV(proaccelerin,labile Serine-typeendopeptidaseactivity 4.24 1.69E-09 8.31E-08 Up-regulated
factor)
FABP5 Fattyacid–bindingprotein5(psoriasis Phosphatidylcholinebiosynthetic 2.66 1.65E-06 4.20E-05 Up-regulated
associated) process
FAM13C Familywithsequencesimilarity13,memberC — 0.20 3.30E-14 3.50E-12 Down-regulated
FAM43A Familywithsequencesimilarity43,memberA — 2.07 6.27E-04 7.65E-03 Up-regulated
FAM46A Familywithsequencesimilarity46,memberA Regulationofbloodcoagulation 2.08 3.85E-08 1.40E-06 Up-regulated
FAM46B Familywithsequencesimilarity46,memberB — 0.45 5.78E-09 2.52E-07 Down-regulated
FAM83D Familywithsequencesimilarity83,memberD Mitoticnucleardivision 0.08 1.17E-17 2.00E-15 Down-regulated
FAR2 FattyacylCoAreductase2 Fatty-acyl-CoAreductase(alcohol- 0.27 1.56E-04 2.36E-03 Down-regulated
forming)activity
FBLN7 Fibulin7 Proteinaceousextracellularmatrix 5.86 5.88E-31 5.03E-28 Up-regulated
FBN2 Fibrillin2 Positiveregulationofosteoblast 6.43 1.52E-04 2.31E-03 Up-regulated
differentiation
FCER1A FcfragmentofIgE,high-affinityI,receptorfor Positiveregulationofgranulocyte- 5.57 2.26E-04 3.23E-03 Up-regulated
(cid:2)polypeptide macrophagecolony-stimulating
factorbiosyntheticprocess
FCGR1B FcfragmentofIgG,highaffinityIb,receptor Antigenprocessingandpresentation 11.13 1.06E-22 3.69E-20 Up-regulated
(CD64) ofexogenouspeptideantigenvia
MHCclassI,TAPdependent
FCGR2A FcfragmentofIgG,lowaffinityIIa,receptor Fc-(cid:3)receptorsignalingpathway 5.14 4.97E-20 1.30E-17 Up-regulated
(CD32) involvedinphagocytosis
FGD2 FYVE,RhoGEFandPHdomaincontaining2 RegulationofsmallGTPase-mediated 18.24 1.95E-09 9.37E-08 Up-regulated
signaltransduction
FGF1 Fibroblastgrowthfactor1(acidic) Positiveregulationoftranscription 5.08 7.70E-23 2.79E-20 Up-regulated
fromRNApolymeraseIIpromoter
FGFR4 Fibroblastgrowthfactorreceptor4 Fibroblastgrowthfactor–activated 2.49 1.88E-05 3.55E-04 Up-regulated
receptoractivity
FHL1 FourandahalfLIMdomains1 Regulationofpotassiumion 0.44 2.13E-07 6.65E-06 Down-regulated
transmembranetransporter
activity
FHL2 FourandahalfLIMdomains2 Negativeregulationoftranscription 4.23 3.18E-22 1.03E-19 Up-regulated
fromRNApolymeraseIIpromoter
FHOD3 Forminhomology2domaincontaining3 Negativeregulationofactinfilament 0.24 2.88E-16 4.11E-14 Down-regulated
polymerization
FLNC FilaminC,(cid:3) Cytoskeletalproteinbinding 0.45 2.89E-07 8.78E-06 Down-regulated
FMNL3 Formin-like3 GTPase-activatingproteinbinding 2.79 3.69E-11 2.47E-09 Up-regulated
FN1 Fibronectin1 Endoplasmicreticulum–Golgi 2.80 3.30E-09 1.53E-07 Up-regulated
intermediatecompartment
FOLR2 Folatereceptor2(fetal) Anchoredcomponentofexternal 4.44 2.72E-10 1.56E-08 Up-regulated
sideofplasmamembrane
Continuedonnextpage
E6 Holcomb ●2015 www.ajnr.org
On-lineTable:Continued
Fold FalseDiscovery Directionof
Gene EntrezGeneName Function Change PValue Rate(qValue) Expression
FOS FBJmurineosteosarcomaviraloncogene Regulationofsequence-specific 2.66 2.11E-06 5.16E-05 Up-regulated
homolog DNA-bindingtranscriptionfactor
activity
FST Follistatin Negativeregulationoftranscription 5.23 2.54E-25 1.26E-22 Up-regulated
fromRNApolymeraseIIpromoter
FYB FYN-bindingprotein NLS-bearingproteinimportinto 7.84 5.02E-05 8.55E-04 Up-regulated
nucleus
GADD45A GrowtharrestandDNA-damageinducible,(cid:2) Regulationofcyclin-dependent 2.11 8.01E-08 2.75E-06 Up-regulated
proteinserine/threoninekinase
activity
GAREM GRB2associated,regulatorofMAPK1 CellularresponsetoEGFstimulus 0.48 4.92E-04 6.18E-03 Down-regulated
GAS2L2 Growtharrest-specific2-like2 Negativeregulationofmicrotubule 0.22 1.46E-04 2.24E-03 Down-regulated
depolymerization
GCNT1 Glucosaminyl(N-acetyl)transferase1,core2 (cid:4)-1,3-Galactosyl-O-glycosyl- 4.46 6.44E-04 7.81E-03 Up-regulated
glycoprotein(cid:4)-1,6-N-
acetylglucosaminyltransferase
activity
GDF6 Growth-differentiationfactor6 Positiveregulationofpathway- 3.30 3.21E-04 4.34E-03 Up-regulated
restrictedSMADprotein
phosphorylation
GGH (cid:3)-Glutamylhydrolase(conjugase, (cid:3)-Glutamyl-peptidaseactivity 3.99 1.90E-06 4.73E-05 Up-regulated
folylpolygammaglutamylhydrolase)
GLB1 Galactosidase,(cid:4)1 Glycosaminoglycancatabolicprocess 2.84 1.78E-13 1.66E-11 Up-regulated
GLDN Gliomedin Proteinbindinginvolvedin 11.50 3.83E-18 7.07E-16 Up-regulated
heterotypiccell–celladhesion
GLP2R Glucagon-likepeptide2receptor Adenylatecyclase-modulating 17.27 7.00E-11 4.39E-09 Up-regulated
G-protein–coupledreceptor
signalingpathway
GMFG Gliamaturationfactor,(cid:3) Negativeregulationofproteinkinase 7.59 3.41E-05 6.13E-04 Up-regulated
activity
GNAO1 Guaninenucleotide-bindingprotein Adenylatecyclase-modulating 0.32 4.31E-09 1.92E-07 Down-regulated
(Gprotein),(cid:2)-activatingactivity G-protein–coupledreceptor
polypeptideO signalingpathway
GNG2 Guaninenucleotide-bindingprotein Adenylatecyclase-activating 3.18 6.50E-05 1.08E-03 Up-regulated
(Gprotein),(cid:3)2 dopaminereceptorsignaling
pathway
GNGT2 Guaninenucleotide-bindingprotein G-protein–coupledreceptorsignaling 4.22 4.35E-05 7.51E-04 Up-regulated
(Gprotein),(cid:3)-transducingactivity pathway
polypeptide2
GPNMB Glycoprotein(transmembrane)nmb Negativeregulationofcell 4.26 4.65E-08 1.66E-06 Up-regulated
proliferation
GPR183 G-protein–coupledreceptor183 MatureB-celldifferentiationinvolved 9.42 9.95E-09 4.02E-07 Up-regulated
inimmuneresponse
GPR34 G-protein–coupledreceptor34 G-protein–coupledreceptorsignaling 8.01 1.59E-18 3.34E-16 Up-regulated
pathway
GPR65 G-protein–coupledreceptor65 PositiveregulationofcAMP 19.44 2.00E-06 4.94E-05 Up-regulated
biosyntheticprocess
GPR97 G-protein–coupledreceptor97 G-protein–coupledreceptoractivity 7.16 6.86E-10 3.67E-08 Up-regulated
GREM1 Gremlin1,DANfamilyBMPantagonist Positiveregulationoftranscription 3.65 1.98E-15 2.49E-13 Up-regulated
fromRNApolymeraseIIpromoter
involvedinmyocardialprecursor
celldifferentiation
GREM2 Gremlin2,DANfamilyBMPantagonist Cytokine-mediatedsignalingpathway 3.39 5.02E-06 1.11E-04 Up-regulated
GRN Granulin Positiveregulationofepithelialcell 2.10 1.02E-06 2.75E-05 Up-regulated
proliferation
HAVCR1 HepatitisAviruscellularreceptor1 Integralcomponentofmembrane 11.58 2.28E-08 8.58E-07 Up-regulated
HCK Hematopoieticcellkinase Regulationofsequence-specific 26.15 2.11E-11 1.46E-09 Up-regulated
DNA-bindingtranscriptionfactor
activity
HCLS1 Hematopoieticcell-specificLynsubstrate1 Negativeregulationoftranscription 14.90 1.37E-09 6.83E-08 Up-regulated
fromRNApolymeraseIIpromoter
HEXA HexosaminidaseA((cid:2)polypeptide) Cellmorphogenesisinvolvedin 2.30 6.41E-09 2.75E-07 Up-regulated
neurondifferentiation
HHATL Hedgehogacyltransferase-like NegativeregulationofN-terminal 0.29 4.69E-04 5.93E-03 Down-regulated
proteinpalmitoylation
HHIP Hedgehog-interactingprotein Oxidoreductaseactivity,actingon 0.28 2.91E-10 1.64E-08 Down-regulated
theCHOOHgroupofdonors,
quinoneorsimilarcompoundas
acceptor
HK3 Hexokinase3(whitecell) Glucose6-phosphatemetabolic 11.49 1.02E-05 2.05E-04 Up-regulated
process
HLA-A MHCclassI,A Antigenprocessingandpresentation 2.71 6.59E-06 1.42E-04 Up-regulated
ofendogenouspeptideantigen
viaMHCclassIviaERpathway,
TAPindependent
HLA-DMA MHCclassII,DM(cid:2) Antigenprocessingandpresentation 26.08 4.57E-14 4.63E-12 Up-regulated
ofexogenouspeptideantigenvia
MHCclassII
HLA-DMB MHCclassII,DM(cid:4) PositiveregulationofT-cell 26.50 3.74E-13 3.35E-11 Up-regulated
activationviaT-cellreceptor
contactwithantigenboundto
MHCmoleculeonantigen-
presentingcell
Continuedonnextpage
AJNRAmJNeuroradiol●:● ●2015 www.ajnr.org E7
On-lineTable:Continued
Fold FalseDiscovery Directionof
Gene EntrezGeneName Function Change PValue Rate(qValue) Expression
HLA-DOB MHCclassII,DO(cid:4) Negativeregulationofantigen 11.08 8.39E-07 2.31E-05 Up-regulated
processingandpresentationof
peptideantigenviaMHCclassII
HLA-DPA1 MHCclassII,DP(cid:2)1 Antigenprocessingandpresentation 10.98 8.71E-08 2.96E-06 Up-regulated
ofexogenouspeptideantigenvia
MHCclassII
HLA-DPB1 MHCclassII,DP(cid:4)1 Antigenprocessingandpresentation 8.04 1.97E-05 3.71E-04 Up-regulated
ofexogenouspeptideantigenvia
MHCclassII
HLA-DQA1 MHCclassII,DQ(cid:2)1 Antigenprocessingandpresentation 46.24 1.33E-19 3.30E-17 Up-regulated
ofexogenouspeptideantigenvia
MHCclassII
HLA-DQB2 MHCclassII,DQ(cid:4)2 Antigenprocessingandpresentation 12.42 2.69E-12 2.17E-10 Up-regulated
ofexogenouspeptideantigenvia
MHCclassII
HLA-DRA MHCclassII,DR(cid:2) Antigenprocessingandpresentation 41.87 2.42E-18 4.75E-16 Up-regulated
ofpeptideorpolysaccharide
antigenviaMHCclassII
HLA-DRB1 MHCclassII,DR(cid:4)1 Positiveregulationofinsulin 44.42 2.50E-18 4.80E-16 Up-regulated
secretioninvolvedincellular
responsetoglucosestimulus
HLA-DRB5 MHCclassII,DR(cid:4)5 Positiveregulationofinsulin 14.96 1.13E-09 5.81E-08 Up-regulated
secretioninvolvedincellular
responsetoglucosestimulus
HMCN1 Hemicentin1 Extracellularvesicularexosome 0.48 4.28E-08 1.54E-06 Down-regulated
HMGCLL1 3-Hydroxymethyl-3-methylglutaryl-CoA Hydroxymethylglutaryl-CoAlyase 0.23 7.21E-06 1.51E-04 Down-regulated
lyase-like1 activity
HMOX1 Hemeoxygenase(decycling)1 RegulationoftranscriptionfromRNA 9.52 7.20E-12 5.43E-10 Up-regulated
polymeraseIIpromoterin
responsetooxidativestress
HNMT HistamineN-methyltransferase HistamineN-methyltransferase 3.22 7.34E-09 3.09E-07 Up-regulated
activity
HP Haptoglobin Negativeregulationofhydrogen 3.00 5.08E-14 5.09E-12 Up-regulated
peroxidecatabolicprocess
HPGD Hydroxyprostaglandindehydrogenase15(NAD) Transforminggrowthfactor(cid:4) 0.20 8.68E-12 6.44E-10 Down-regulated
receptorsignalingpathway
HPSE Heparanase Positiveregulationvascular 3.66 5.44E-05 9.20E-04 Up-regulated
endothelialgrowthfactor
production
HPSE2 Heparanase2 Heparansulfateproteoglycanbinding 0.38 3.26E-04 4.39E-03 Down-regulated
HRH1 HistaminereceptorH1 PhospholipaseC-activating 0.31 2.03E-09 9.72E-08 Down-regulated
G-protein–coupledreceptor
signalingpathway
HS3ST1 Heparansulfate(glucosamine) (cid:3)heparansulfate(cid:4)-Glucosamine3- 0.45 3.73E-04 4.92E-03 Down-regulated
3-O-sulfotransferase1 sulfotransferase1activity
HSPA1A/HSPA1B Heatshock70-kDaprotein1A — 0.44 1.16E-07 3.89E-06 Down-regulated
HSPA1A/HSPA1B Heatshock70-kDaprotein1A — 0.42 1.02E-05 2.05E-04 Down-regulated
HSPA1A/HSPA1B Heatshock70kDaprotein1A — 0.41 9.44E-10 4.91E-08 Down-regulated
HTRA1 HtrAserinepeptidase1 Negativeregulationoftransforming 2.11 3.75E-07 1.13E-05 Up-regulated
growthfactor(cid:4)receptorsignaling
pathway
IFIT3 Interferon-inducedproteinwith Negativeregulationofcell 2.82 2.41E-05 4.47E-04 Up-regulated
tetratricopeptiderepeats3 proliferation
IGFALS Insulin-likegrowthfactor-bindingprotein, Cellularproteinmetabolicprocess 5.93 9.25E-05 1.49E-03 Up-regulated
acid-labilesubunit
IGJ IgJpolypeptide,linkerproteinforIg(cid:2)and Positiveregulationofprotein 4.93 2.92E-04 4.01E-03 Up-regulated
Ig(cid:7)polypeptides oligomerization
IGSF3 Igsuperfamily,member3 Integralcomponentofmembrane 0.48 2.46E-07 7.53E-06 Down-regulated
IGSF6 Igsuperfamily,member6 Transmembranesignalingreceptor 12.07 3.04E-16 4.27E-14 Up-regulated
activity
IL13RA2 Interleukin13receptor,(cid:2)2 Cytokine-mediatedsignalingpathway 4.28 8.25E-07 2.29E-05 Up-regulated
IL15 Interleukin15 Positiveregulationoftyrosine 3.91 4.11E-04 5.33E-03 Up-regulated
phosphorylationofStat3protein
IL1B Interleukin1(cid:4) Positiveregulationofvascular 23.09 1.70E-08 6.52E-07 Up-regulated
endothelialgrowthfactor
receptorsignalingpathway
IL1R1 Interleukin1receptortypeI Interleukin1,typeI,activating 2.09 1.77E-07 5.69E-06 Up-regulated
receptoractivity
IL1RL2 Interleukin1receptor-like2 Interleukin-1,typeI,activating 7.24 7.60E-08 2.64E-06 Up-regulated
receptoractivity
IL1RN Interleukin1receptorantagonist Negativeregulationofinterleukin 24.95 1.40E-10 8.32E-09 Up-regulated
1–mediatedsignalingpathway
IL21R Interleukin21receptor Interleukin21–mediatedsignaling 24.86 2.36E-06 5.69E-05 Up-regulated
pathway
IL3RA Interleukin3receptor,(cid:2)(lowaffinity) Interleukin3–mediatedsignaling 4.80 5.73E-04 7.09E-03 Up-regulated
pathway
IL7 Interleukin7 Negativeregulationofextrinsic 3.61 1.49E-04 2.27E-03 Up-regulated
apoptoticsignalingpathwayin
absenceofligand
INPP4A Inositolpolyphosphate-4-phosphatase,typeI, Phosphatidylinositol-3,4- 0.49 1.87E-07 5.96E-06 Down-regulated
107kDa bisphosphate4-phosphatase
activity
IRF5 Interferonregulatoryfactor5 Positiveregulationoftranscription 18.44 1.42E-15 1.81E-13 Up-regulated
fromRNApolymeraseIIpromoter
Continuedonnextpage
E8 Holcomb ●2015 www.ajnr.org
On-lineTable:Continued
Fold FalseDiscovery Directionof
Gene EntrezGeneName Function Change PValue Rate(qValue) Expression
IRG1 Immunoresponsive1homolog(mouse) Positiveregulationofreactiveoxygen 9.95 3.75E-05 6.61E-04 Up-regulated
speciesmetabolicprocess
IRGC Immunity-relatedGTPasefamily,cinema Hydrolaseactivity,actingonacid 4.34 3.18E-04 4.31E-03 Up-regulated
anhydrides
IRX5 Iroquoishomeobox5 Sequence-specificDNA-binding 11.81 4.03E-09 1.82E-07 Up-regulated
transcriptionfactoractivity
IRX6 Iroquoishomeobox6 Sequence-specificDNA-binding 3.38 3.56E-04 4.74E-03 Up-regulated
transcriptionfactoractivity
ITGA10 Integrin,(cid:2)10 Integrin-mediatedsignalingpathway 3.51 4.98E-19 1.14E-16 Up-regulated
ITGA2B Integrin,(cid:2)2b(plateletglycoproteinIIbof Positiveregulationofleukocyte 0.33 5.64E-06 1.24E-04 Down-regulated
IIb/IIIacomplex,antigenCD41) migration
ITGA3 Integrin,(cid:2)3(antigenCD49C,(cid:2)3subunitof Positiveregulationofneuron 2.67 8.05E-08 2.76E-06 Up-regulated
VLA-3receptor) projectiondevelopment
ITGA5 Integrin,(cid:2)5(fibronectinreceptor, Positiveregulationofvascular 0.43 1.59E-08 6.13E-07 Down-regulated
(cid:2)polypeptide) endothelialgrowthfactor
receptorsignalingpathway
ITGBL1 Integrin,(cid:4)-like1(withEGF-likerepeatdomains) Extracellularregion 2.06 8.84E-06 1.80E-04 Up-regulated
ITIH3 Inter-(cid:2)-trypsininhibitorheavy-chain3 Negativeregulationof 5.61 4.99E-13 4.43E-11 Up-regulated
endopeptidaseactivity
KANK4 KNmotifandankyrinrepeatdomains4 Cytoplasm 7.44 2.67E-27 1.68E-24 Up-regulated
KCNAB1 Potassiumvoltage-gatedchannel,shaker- Regulationofpotassiumion 0.25 6.83E-19 1.53E-16 Down-regulated
relatedsubfamily,(cid:4)member1 transmembranetransporter
activity
KCNH1 Potassiumvoltage-gatedchannel,subfamilyH Delayedrectifierpotassiumchannel 0.24 9.79E-07 2.66E-05 Down-regulated
(eag-related),member1 activity
KCNK3 Potassiumchannel,subfamilyK,member3 Negativeregulationofcytosolic 0.45 3.43E-05 6.14E-04 Down-regulated
calciumionconcentration
KCNQ4 Potassiumvoltage-gatedchannel,KQT-like Delayedrectifierpotassiumchannel 0.41 1.81E-05 3.42E-04 Down-regulated
subfamily,member4 activity
KCNQ5 Potassiumvoltage-gatedchannel,KQT-like Delayedrectifierpotassiumchannel 2.08 2.17E-04 3.11E-03 Up-regulated
subfamily,member5 activity
KCTD12 Potassiumchanneltetramerizationdomain Extracellularvesicularexosome 2.70 2.67E-09 1.26E-07 Up-regulated
containing12
KDR Kinaseinsertdomainreceptor(atypeIII Positiveregulationofnitric-oxide 3.25 1.18E-09 6.00E-08 Up-regulated
receptortyrosinekinase) synthasebiosyntheticprocess
KIAA1598 KIAA1598 Kinasebinding 3.29 2.99E-05 5.44E-04 Up-regulated
KIF21B Kinesinfamilymember21B Microtubulemotoractivity 7.76 4.04E-05 7.07E-04 Up-regulated
KIF5C Kinesinfamilymember5C Microtubulemotoractivity 0.42 3.32E-06 7.73E-05 Down-regulated
KIRREL3 KinofIRRE-like3(Drosophila) Principalsensorynucleusof 0.44 4.47E-04 5.71E-03 Down-regulated
trigeminalnervedevelopment
KIT v-kitHardy-Zuckerman4felinesarcomaviral Positiveregulationofsequence- 2.85 1.77E-04 2.64E-03 Up-regulated
oncogenehomolog specificDNA-binding
transcriptionfactoractivity
KLHL40 Kelch-likefamilymember40 Multicellularorganismaldevelopment 0.23 7.91E-06 1.64E-04 Down-regulated
KLHL6 Kelch-likefamilymember6 B-cellreceptorsignalingpathway 7.70 1.66E-04 2.50E-03 Up-regulated
LAPTM5 Lysosomalproteintransmembrane5 Integralcomponentofplasma 9.60 5.95E-13 5.15E-11 Up-regulated
membrane
LCP2 Lymphocytecytosolicprotein2(SH2domain Transmembranereceptorprotein 12.68 3.91E-06 8.93E-05 Up-regulated
containingleukocyteproteinof76kDa) tyrosinekinasesignalingpathway
LECT1 Leukocyte-cell–derivedchemotaxin1 Negativeregulationofvascular 0.31 1.87E-08 7.11E-07 Down-regulated
endothelialgrowthfactor
receptorsignalingpathway
LEF1 Lymphoid-enhancer–bindingfactor1 RNApolymeraseIItranscription 11.03 2.13E-07 6.65E-06 Up-regulated
regulatoryregionsequence-
specificDNA-binding
transcriptionfactoractivity
involvedinpositiveregulationof
transcription
LGALS9B Lectin,galactosidebinding,soluble9B Carbohydratebinding 79.40 3.66E-09 1.67E-07 Up-regulated
LGMN Legumain Antigenprocessingandpresentation 2.22 4.14E-09 1.86E-07 Up-regulated
ofexogenouspeptideantigenvia
MHCclassII
LIPA LipaseA,lysosomalacid,cholesterolesterase Homeostasisofnumberofcells 2.63 1.61E-11 1.13E-09 Up-regulated
withinatissue
LMO2 LIMdomainonly2(rhombotin-like1) RNApolymeraseIItranscription 2.13 1.41E-05 2.74E-04 Up-regulated
regulatoryregionsequence-
specificDNA-binding
transcriptionfactoractivity
involvedinpositiveregulationof
transcription
LNP1 LeukemiaNUP98fusionpartner1 — 0.24 3.75E-05 6.61E-04 Down-regulated
LPCAT2 Lysophosphatidylcholineacyltransferase2 1-Alkylglycerophosphocholine 6.26 1.02E-11 7.38E-10 Up-regulated
O-acetyltransferaseactivity
LPXN Leupaxin NegativeregulationofB-cell 6.17 4.73E-05 8.10E-04 Up-regulated
receptorsignalingpathway
LRRTM3 Leucine-richrepeattransmembraneneuronal3 Positiveregulationof(cid:4)-amyloid 3.85 1.82E-04 2.68E-03 Up-regulated
formation
LY86 Lymphocyteantigen86 Positiveregulationof 18.71 5.28E-09 2.31E-07 Up-regulated
lipopolysaccharide-mediated
signalingpathway
LYN v-yes-1Yamaguchisarcomaviral-related PositiveregulationofFcreceptor- 5.48 1.11E-13 1.06E-11 Up-regulated
oncogenehomolog mediatedstimulatorysignaling
pathway
Continuedonnextpage
AJNRAmJNeuroradiol●:● ●2015 www.ajnr.org E9
On-lineTable:Continued
Fold FalseDiscovery Directionof
Gene EntrezGeneName Function Change PValue Rate(qValue) Expression
LYPD6 LY6/PLAURdomain-containing6 Extracellularregion 0.44 5.81E-06 1.27E-04 Down-regulated
LYZ Lysozyme Cellwallmacromoleculecatabolic 10.44 1.16E-08 4.60E-07 Up-regulated
process
LZTS1 Leucinezipper,putativetumor Sequence-specificDNA-binding 2.10 8.45E-04 9.88E-03 Up-regulated
suppressor1 transcriptionfactoractivity
MAL2 Mal,T-celldifferentiation Extracellularvesicularexosome 0.11 2.65E-08 9.91E-07 Down-regulated
protein2(gene/
pseudogene)
MAN1C1 Mannosidase,(cid:2),class1C, Mannosyl-oligosaccharide1,2-(cid:2)- 2.05 8.13E-06 1.67E-04 Up-regulated
member1 mannosidaseactivity
MAP4K1 MAPKkinasekinasekinase1 MAPKkinasekinasekinaseactivity 6.25 4.73E-04 5.97E-03 Up-regulated
MAPK4 MAPK4 Proteinheterodimerizationactivity 0.27 7.85E-17 1.21E-14 Down-regulated
MARCKS Myristoylatedalanine-rich Energy-reservemetabolicprocess 2.15 4.29E-05 7.47E-04 Up-regulated
proteinkinaseCsubstrate
MARCO Macrophagereceptorwith Signalingpattern-recognition 45.60 4.33E-09 1.93E-07 Up-regulated
collagenousstructure receptoractivity
MATN3 Matrilin3 Extracellularmatrixstructural 35.90 2.59E-18 4.88E-16 Up-regulated
constituent
MB21D1 Mab-21domaincontaining1 Positiveregulationofdefense 7.26 2.92E-06 6.89E-05 Up-regulated
responsetovirusbyhost
MCOLN2 Mucolipin2 Calciumiontransmembrane 23.65 3.00E-08 1.11E-06 Up-regulated
transport
ME3 Malicenzyme3,NADP((cid:2)) Malatedehydrogenase 0.33 8.31E-11 5.12E-09 Down-regulated
dependent,mitochondrial (decarboxylating)(NADP(cid:2))
activity
MEGF10 MultipleEGF-likedomains10 Regulationofskeletalmuscletissue 2.48 5.60E-04 6.96E-03 Up-regulated
development
MEIS2 Meishomeobox2 RNApolymeraseIIcorepromoter 0.36 5.94E-09 2.58E-07 Down-regulated
proximalregionsequence-specific
DNA-bindingtranscriptionfactor
activityinvolvedinpositive
regulationoftranscription
MFAP3L Microfibrillar-associated Integralcomponentofmembrane 0.32 7.80E-10 4.13E-08 Down-regulated
protein3-like
MFAP5 Microfibrillar-associated Extracellularmatrixstructural 0.37 1.35E-08 5.29E-07 Down-regulated
protein5 constituent
MFI2 Antigenp97(melanoma Negativeregulationofsubstrate 12.28 5.50E-10 2.98E-08 Up-regulated
associated)identifiedby adhesion-dependentcell
monoclonalantibodies133.2 spreading
and96.5
MGP MatrixGlaprotein Extracellularmatrixstructural 4.74 1.80E-18 3.70E-16 Up-regulated
constituent
MILR1 MastcellIg-likereceptor1 Negativeregulationofmastcell 15.14 1.46E-07 4.75E-06 Up-regulated
activation
miR-1(andothermiRNAswith — 0.48 4.62E-05 1.01E-03 Down-regulated
seedGGAAUGU)
miR-10a-5p(andothermiRNAs — 2.66 8.83E-09 4.66E-07 Up-regulated
withseedACCCUGU)
miR-10a-5p(andothermiRNAs — 2.45 3.47E-07 1.47E-05 Up-regulated
withseedACCCUGU)
miR-146a-5p(andothermiRNAs — 3.32 9.08E-11 6.39E-09 Up-regulated
withseedGAGAACU)
miR-146a-5p(andothermiRNAs — 3.68 1.45E-06 4.38E-05 Up-regulated
withseedGAGAACU)
miR-204-5p(andothermiRNAs — 0.48 7.32E-06 1.93E-04 Down-regulated
withseedUCCCUUU)
miR-21-5p(andothermiRNAs — 3.51 4.43E-11 4.67E-09 Up-regulated
withseedAGCUUAU)
miR-223-3p(miRNAswithseed — 3.95 6.79E-12 1.43E-09 Up-regulated
GUCAGUU)
miR-34a-5p(andothermiRNAs — 3.79 1.28E-06 4.38E-05 Up-regulated
withseedGGCAGUG)
miR-34a-5p(andothermiRNAs — 2.34 1.15E-04 2.02E-03 Up-regulated
withseedGGCAGUG)
miR-9-5p(andothermiRNAs — 0.43 4.80E-05 1.01E-03 Down-regulated
withseedCUUUGGU)
MLANA Melan-A Integralcomponentofplasma 13.86 1.25E-09 6.32E-08 Up-regulated
membrane
MMP12 Matrixmetallopeptidase12 Positiveregulationofepithelialcell 44.11 2.61E-17 4.23E-15 Up-regulated
(macrophageelastase) proliferationinvolvedinwound
healing
MMRN1 Multimerin1 Platelet(cid:2)granulelumen 3.94 9.16E-09 3.77E-07 Up-regulated
MPEG1 Macrophageexpressed1 Integralcomponentofmembrane 17.10 1.16E-14 1.35E-12 Up-regulated
MRC1 Mannosereceptor,Ctype1 Integralcomponentofplasma 2.55 1.86E-09 9.02E-08 Up-regulated
membrane
MS4A4A Membrane-spanning4domains, Integralcomponentofmembrane 3.67 2.36E-08 8.86E-07 Up-regulated
subfamilyA,member4A
MS4A6A Membrane-spanning4-domains, Integralcomponentofmembrane 4.63 2.66E-11 1.80E-09 Up-regulated
subfamilyA,member6A
MSR1 Macrophagescavenger Positiveregulationofmacrophage- 28.78 1.13E-29 8.20E-27 Up-regulated
receptor1 derivedfoamcelldifferentiation
Continuedonnextpage
E10 Holcomb ●2015 www.ajnr.org
Description:Atypical chemokine receptor 1 (Duffy blood Down-regulated Ankyrin repeat domain 1 (cardiac muscle) .. Positive regulation of myoblast growth factor receptor signaling .. motility. 0.09. 1.52E-18. 3.25E-16. Down-regulated. DNAJB5. DnaJ (Hsp40) homolog Mannose receptor, C type 1.