Table Of ContentStructure
Article
The NMDA Receptor NR1 C1 Region Bound
to Calmodulin: Structural Insights into Functional
Differences between Homologous Domains
ZeynepAkyolAtaman,1LokeshGakhar,1BrendaR.Sorensen,1JohannesW.Hell,2andMadelineA.Shea1,*
1DepartmentofBiochemistry
2DepartmentofPharmacology
RoyJ.andLucilleA.CarverCollegeofMedicine,UniversityofIowa,IowaCity,IA52242-1109,USA
*Correspondence:[email protected]
DOI10.1016/j.str.2007.10.012
SUMMARY includingcalmodulin(CaM).CaMhastwohighlyhomolo-
gousdomains(NandC);eachdomainhastwoEF-hand
Calmodulin (CaM) regulates tetrameric N- sites that bind calcium cooperatively. CaM regulates
methyl-D-aspartate receptors (NMDARs) by a widearray of target proteins, including kinases, phos-
binding tightly to the C0 and C1 regions of its phatases, and ion channels (cf. Bhattacharya et al.,
NR1subunit.Acrystalstructure(2HQW;1.96A˚) 2004;ChinandMeans,2000;SaimiandKung,2002;Vet-
of calcium-saturated CaM bound to NR1C1 terandLeclerc,2003).
Upon calcium influx, CaM induces inactivation of
(peptide spanning 875–898) showed that NR1
NMDAR(reducingitsopenrateandmeanopentime)by
S890, whose phosphorylation regulates mem-
bindingtoNR1C0andC1(Ehlersetal.,1996b).Inactiva-
brane localization, was solvent protected,
tionreleasesNMDARfromtheneuronalcytoskeletonby
whereas the endoplasmic reticulum retention
disrupting interactions between NR1 and a-actinin2
motifwassolventexposed.NR1F880filledthe
(Krupp et al., 1999; Zhang et al., 1998). CaM binding to
CaMC-domainpocket,whereasT886wasclos- NR1C1significantlyenhancesNMDARinactivation(Eh-
est to the N-domain pocket. This 1-7 pattern lers et al., 1996b), which absolutely requires C0 (Zhang
was most similar to that in the CaM-MARCKS et al., 1998). Dissociation constants for (Ca2+) -CaM
4
complex. Comparison of CaM-ligand wrap- binding to peptides representing the CaM-binding do-
around conformations identified a core tetrad mains (CaMBDs) of NR1 showed that its affinity for C1
of CaM C-domain residues (FLMM ) that con- (NR1C1p; aa 875–898) was 20-fold more favorable than
C
its affinity for C0 (NR1C0p; aa 838–863) (Ehlers et al.,
tacted all ligands consistently. An identical
1996b).TheK values(4and87nM,respectively)suggest
tetradofN-domainresidues(FLMM )madevar- D
N that (Ca2+) -CaM binds both C0 and C1 intracellularly,
iable sets of contacts with ligands. This CaM- 4
where [CaM] is 50–75 nM (Wu and Bers, 2007). C1
free
NR1C1 structure provides a foundation for
hasanendoplasmicreticulum(ER)retentionmotif(R893–
designing mutants to test the role of CaM in
R895;Figure1A)andaProteinKinaseC(PKC)phosphor-
NR1traffickingaswellasinsightsintohowthe ylationsite(S896)thatareneededforpropermaturation
homologousCaMdomainshavedifferentroles andERrelease(Scottetal.,2001;Standleyetal.,2000).
inmolecularrecognition. NR1 S890 phosphorylation by PKC disrupts surface-
associatedclustersofNR1,causinganevendistribution
throughout fibroblasts (Tingley et al., 1997), and affects
INTRODUCTION receptorpotentiation(Zhengetal.,1999).Thepositionof
CaM relative to these regulatory motifs in NR1 is not
The ionotropic N-methyl-D-aspartate receptor (NMDAR) known.
isamajorsourceofcalciumfluxintoneuronsinthebrain ToexploretheroleofCaMinNR1trafficking,itisnot
andhasacriticalroleinlearning,memory,neuraldevelop- possible to make a viable CaM knockout organism be-
ment, and synaptic plasticity (Mori and Mishina, 1995). causeCaMisessentialandhasmultipletargets.Ourstrat-
MammalianNMDARshavetwofamiliesofsubunitsdesig- egy was to determine the binding interface to provide
natedNMDAR1(NR1)andNMDAR2(NR2).Understand- aplatformfortargetedmutagenesisofNR1.Here,were-
ing the roles of NR1 and NR2 subunits in brain function portacrystalstructureof(Ca2+) -CaMboundtoNR1C1p
4
is complicated by variable developmental and spatial (1.96 A˚ resolution) in which the CaM domains wrapped
expressionof theirmRNA,andbythepresence ofeight aroundhelicalNR1C1p(Figures2Aand2B).TheERreten-
variants of NR1 arising from N-terminal sequence varia- tionmotif(R893–R895)andS896weresolventexposed,
tionsandalternativesplicingoffourexonsencodingthe whereas S890 was buried in the N domain of CaM.
C0 (membrane-proximal), C1, C2, and C20 regions (Fig- NR1C1pwaspredictedtobea1-12CaMBD(Yapetal.,
ure1A).TheC-terminaltailofNR1bindsseveralproteins, 2000), but it was found to be a 1-7 CaMBD. NR1 F880
Structure15,1603–1617,December2007ª2007ElsevierLtdAllrightsreserved 1603
Structure
CalmodulinBoundtoNMDARNR1C1Region
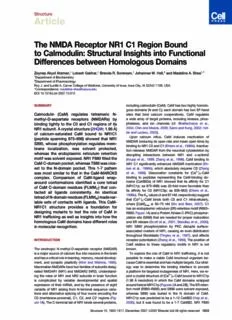
Figure2. CrystalStructureoftheCaM-NR1C1pComplex
(A) NR1C1p sequence and structure superimposed on its electron
densitymapcontouredat1.0s.
(BandC)AlternateviewsofCaM-NR1C1p(2HQW)showingtheCaM
N-domainbackbone(blue),theCdomain(red),Ca2+ionsandbinding
sites(yellow),andNR1C1p(gray).ThefigurewasmadewithMacPymol.
(DandE)Alignmentof17canonicalCaM-targetcomplexesbytheirCa
atomsofthe(D)N-domain(residues5–72;68atoms)and(E)C-domain
(residues84–146;63atoms)FLMMresiduesasdescribedinExperi-
mentalProcedures.
Figure1. CaMBindingtoNMDARNR1
(A)SchematicdiagramofNR1indicatingrelativepositionsofintracel- regulatingMARCKSattachmenttothecytoskeletonand
lularregionsC0,C1,andC2/C20,andsequencesofC0andC1.The
proteinsbindingC1suggestasimilarmechanismofregu-
CaMBDsequenceofC0(residues838–865)showsthesingletrypto-
lationofC1intheformationofNR1-richclusters(Ehlers
phan(presumedanchor)residueboxed.ThesequenceofC1(residues
875–898)isshownwiththeERretentionsignal(RRR)underlined,the etal.,1996b;Tingleyetal.,1997).
PKCsitesboxedandshaded,andresiduesF880andT886boxed. The1-7motiffoundhereisunusualamongCaM-target
(B)BindingofCaMtoNR1C1pmonitoredbyfluorescenceanisotropy interfacesin17complexesinwhichboththeNandCdo-
ofFl-NR1C1p(intrinsicvalueof0.04)toafinalconcentrationof51.5mM mainsofCaMcontactsthetarget.Theonlyotherknown
apoCaM(open;K =158mM)or0.76mM(Ca2+)-CaM(filled;K =
D 4 D case is CaM bound to MARCKS. To determine whether
1.99 nM). The asterisk indicates that the anisotropy of Fl-NR1C1p
thenatureaswellasthespacingofCaMresiduescritical
titratedwithapoCaMwasnormalizedtothevalue(0.13)observed
aftersaturationwithcalcium. to molecular recognition were different among these
(C) Simulation of apo CaM (dashed black) and (Ca2+)-CaM (solid structures, we analyzed the CaM-target contacts in all
4
black) binding to NR1C1p with equilibrium constants from (B). For 17complexesandfoundthat4residuesintheCdomain
comparison,bindingofCaMtoNR1C0p(gray)simulatedwithaKD (F92,L105,M124,andM144:FLMMC)wereusedconsis-
of87nM(Ehlersetal.,1996b)for(Ca2+)4-CaM(solid)andaKDof tentlybyCaMtocontacttargets.Althoughastructurally
2.25mM(Akyoletal.,2004)forapoCaM(dashed).
equivalenttetrad(F19,L32,M51,andM71:FLMM )was
N
was anchored in the C domain of CaM, whereas T886 observed in the CaM N domain, these CaM residues
(ratherthanF891)contactedthehighestnumberofresi- werenotusedidenticallybyalltargets.
duesintheCaMNdomain.Thesame1-7motifandnearly
identicalprimarycontactresidueswereidentifiedforCaM RESULTS
bound to a MARCKS (myristoylated, alanine-rich, PKC
substrate)peptide(1IWQ).Inthatcase,CaMbindinginter- BindingofCaMtoNR1C1pandNR1C0p
ruptsattachmentoftheactincytoskeletontotheplasma TitrationsofNR1C1pwith(Ca2+) -CaMandapo(Ca2+-de-
4
membrane (Aderem, 1992). Parallels between proteins pleted)CaM(Figure1B)yieldedaK of2.0±0.1nMfor
D
1604 Structure15,1603–1617,December2007ª2007ElsevierLtdAllrightsreserved
Structure
CalmodulinBoundtoNMDARNR1C1Region
(Ca2+) -CaM, which agreed well with the value of 4 nM
4 Table1. CrystallographicDataCollectionand
determinedpreviously(Ehlersetal.,1996b).AKDof158± RefinementStatistics
3mM(K of6.33E3M(cid:2)1)wasresolvedforapoCaMbind-
A (Ca2+) -CaM-NRC1pCrystal
ingtoNR1C1p.ComparedtoNR1C0,(Ca2+) -CaMbinds 4
4
DataCollectionStatistics
C0withaK of87nM(Ehlersetal.,1996b)andapoCaM
D
bindsC0withaKDof2.25mM(Akyoletal.,2004).Compar- Spacegroup P3221
isonofsimulatedequilibriumtitrationsofCaMbindingto Celldimensions a=40.361A˚;b=40.361A˚;
NR1C1p andNR1C0p(Figure1C)showedthatthemid- c=175.765A˚;a=90(cid:3);
pointsforapoand(Ca2+) -CaMbindingtoNR1Cpdiffered b=90(cid:3);g=120(cid:3)
4
byfourordersofmagnitude. Resolution(A˚) 19.69(cid:2)1.90(2.00(cid:2)1.90)
Structureofthe(Ca2+) -CaM-NR1C1pComplex RsymorRmerge 0.034(0.255)
4
The crystal structure of (Ca2+) -CaM bound to NR1C1p I/sI 17.36(3.92)
4
was determined to 1.96 A˚ resolution (Figures 2A–2C). It
Completeness(%) 98.4(92.6)
adoptedthecanonicalCaM-targetconformationinwhich
Redundancy 3.01(2.56)
boththeNandCdomainsofCaMcontactedpeptideto
formacompact,ellipsoidalcomplex.CaMresidues1,2, RefinementStatistics
75–80, and 148 were disordered and were not included Resolution(A˚) 8.57(cid:2)1.90
inthemodel.Figure2Ashowstheelectrondensitymap
Numberofreflections 13,379
for NR1C1p (residues S897 and K898 were disordered);
thepeptidewasbuiltintothefinalmodelmanually.Refine- Rwork/Rfree(%) 20.6/24.8
mentstatisticsaregiveninTable1. Bfactorforprotein(A˚) 33.8
This structure was compared to 16 other similar and
Bfactorforligand 43.5
nonredundant(Ca2+) -CaM-targetstructures(listedinEx-
4
perimentalProcedures).Toevaluatetheoverallstructural Bfactorforions 31.4
variabilityinthese17compactCaM-targetcomplexes,we Bfactorforwater 41.8
comparedeachonetoanaveragestructureasdescribed
Rmsdbondlengths(A˚) 0.019
inExperimentalProcedures.Among17structures,theav-
eragermsdoftheCaMNdomainwas0.75A˚,whereasthat Rmsdbondangles((cid:3)) 1.585
oftheCdomainwas0.59A˚ (Figures2Dand2E).Thecom- Numberofproteinatoms 1,089
plexesthatshowedthehighestdeviationfromtheaverage
Numberofligandatoms 178
backboneconformationintheCdomainwerethosede-
terminedbyNMR(CaMwithCNGchannel[1SY9],CaMKK Numberofions 4
[1CKK],andskMLCK[2BBM]);skMLCKalsohadthehigh- Numberofwateratoms 66
estN-domaindeviation. Twostructures ofthedrugTFP
Ramachandranplot(%residues)
bound to CaM (1A29 for the N domain; 1LIN for the C
domain)hadthesmallestrmsdvalues. Mostfavored 94.2
Additionallyallowed 5.8
AccessibilityofNR1C1pMotifs
Disallowed 0.0
Processing and localization of C1-containing NR1 sub-
units is regulated by an ER retention motif (R893–R895) Valuesinparenthesesrefertothehighest-resolutionshell.
and by phosphorylation of S896. These residues had
ahighfractionalsolvent-accessiblesurfacearea(SASA):
R893,89.4%;R894,71.6%;R895,92.4%;S896,87.1%; (Figure 4). Among the ordered side chains, this analysis
averageSASA,85%(Figure3A).Incontrast,S890(impli- identified 36 residues in the N domain (residues 3–74)
cated in subunit clustering and receptor potentiation by and34intheCdomain(residues81–147)thatmetthiscri-
PKC) was protected by the N domain of CaM, having terion.AsshowninFigures4Aand4B,contactswiththe
only 41% SASA. Two CaM N-domain residues (M36, CaMNdomainwerewelldistributedacrossthelengthof
M51)werewithin4.5A˚ ofNR1S890asdeterminedbyus- NR1C1p: 17 with the N-terminal half (residues 875–885;
ing Contacts of Structural Units (CSU) (Sobolev et al., gray)and19withtheC-terminalhalf(residues886–896;
1999). The hydroxyl of S890 was 3.22 A˚ from the sulfur black). In contrast, contacts with the CaM C domain
ofM36and4.01A˚ fromthatofM51,suggestingthatthese were skewed: 27 with the N-terminal half of NR1C1p
residuesinteractinthecomplex(Figure3B).Nocontacts andonly7withtheC-terminalhalf(Figures4Band4C).
were observed between NR1 S890 and any residues in While CSU analysis showed that most NR1C1p resi-
theCaMCdomain. dues contacted a single CaM domain, side chains of
NR1C1p K875, K876, T879, and L887 contacted 2 or
InterfaceContacts more residues in each domain. The 7 contacts of K875
ToexploretheCaM-NR1C1pinterface,CSUwasusedto are shown in purple and are underlined in Figure 5A.
determineCaMresidueswithin4.5A˚ ofNR1C1presidues NR1C1pF880contactedthehighestnumberofresidues
Structure15,1603–1617,December2007ª2007ElsevierLtdAllrightsreserved 1605
Structure
CalmodulinBoundtoNMDARNR1C1Region
main(i.e.,FLMM andFLMM )appearedtoadoptnearly
N C
identical spatial conformations (Figure 5C). The electron
density overlap of side chains of FLMM residues and
C
NR1C1pF880isshowninFigure5D.Theperpendicular
orientation of NR1C1p F880 relative to CaM F92 allows
forafavorablep-pinteractionbetweenthetwoaromatic
rings(SinghandThornton,1992).
IdentifyingCaMResiduesCommonlyUsed
forTargetInteractions
ThereisonlyoneotherstructureofCaMboundtoa1-7
CaMBDmotif,buttherearenumerouscompact,ellipsoi-
dal CaM-target structures. To explore how the CaM-
NR1C1p interface related to those complexes, we used
CSU to conduct a statistical analysis of CaM residues
contactingtargetsin16othercompact(Ca2+) -CaM-tar-
4
getstructures(12CaM-peptide,4CaM-drugcomplexes;
listedinExperimentalProcedures).Inthesetof17struc-
tures analyzed, 3 CaM residues (F92, M124, and M144)
contacted every target; in all but one structure, L105
alsocontactedthetarget(Figure6A).Thus,FLMM con-
C
sistentlyservesastheverticesoftheC-domainhydropho-
bic pocket in these structures. An overlay of domains
alignedaccordingtotheCaatomsoftheFLMM tetrad
C
shows that the positions of these FLMM residues in all
structuresisinFigure6B.
AcorrespondinganalysisoftheNdomainshowedthat
Figure3. SolventAccessibilityofS890 althoughresiduesintheFLMM tetradwerecontactedin
N
(A)Surface(CaM)andstick(NR1C1p)diagramof2HQWcoloredas
atleast12oftheanalyzedstructures,theseresidueswere
agradientfromblue(buried)tored(exposed)accordingto%SASA
notthe4residuescontactedmostfrequently(Figure6C).
values:S890,41%;R893,89%;R894,72%;R895,92%;andS896,
87%. Instead,E11wastheonlyresiduefoundtobewithin4.5A˚
(B)Ball-and-stickdiagramofS890andCaMN-domainresidues(M36, of the target peptide or drug in all 17 of the structures
M51,andQ41). that were examined. However, 2 of those 17 structures
ThefigurewasmadewithMacPymol. (1CTR.pdband1A29.pdb)wereCaM-drugcomplexesin
which the pocket of the N domain was vacant. Thus,
(7)withinasingledomainofCaM(Figure4B).F880was E11 interacted with target molecules bound exclusively
within 4.5 A˚ of F92, I100, L105, M124, A128, F141, and in the hydrophobic cleft of the C domain. The second
M144 (red letters in Figure 5A). All have hydrophobic mostcommonlyusedN-domainresidue,A15,alsocon-
sidechainslocatedintheCdomainofCaM.Thepeptide tactsthetargetassociatedwiththeCdomainofCaM.In
residue making the second highest number of contacts the15structuresthathadthehydrophobiccleftoftheN
withinasingledomainofCaMwasT886.Itspartnersin domain occupied, F19 contacted the target in all of
CaMwereF19,L32,M36,M51,andM72,allhydrophobic them,asdidE14.However,thefrequencyofuseofother
sidechainsintheNdomain(bluelettersinFigure5A).In FLMM residues(L32,M51,andM71)waslowerandwas
N
thecommonparlanceofCaM-peptideinteractions,F880 dispersed amongotherhydrophobic N-domainresidues
andT886qualifyaspeptideanchorsinthehydrophobic (L18,M72,M36,F68,andL39)thatcontactedthetarget
clefts of the C and N domains, respectively; however, inasmanyormorestructures.Anoverlayof17Ndomains
the anchors are usually both hydrophobic. Alignment of aligned according to Ca atoms of the FLMM tetrad
N
theCaMsequencebyitscalcium-bindingsites(Figure5A) (Figure6D)showsthatthepocketformedisverysimilar
illustrated that a tetrad of the CaM residues contacting inallstructures.
F880andT886residueswereasetofidenticalsidechains AcomparisonofsidechainorientationsofeachFLMM
(FLMM) in corresponding positions: F19/F92, L32/L105, residueinthese17structuresisshowninFigure6E.Each
M51/M124,andM71/M144(boxed,Figure5A). FLMM residue was aligned with the corresponding resi-
The domains of CaM in complex with NR1C1p were due in 2HQW; rmsds ranged from 0.2 to >1.6 A˚. All
alignedbyminimizingthedistancebetweentheCaatoms FLMMresidues,exceptF92,hadsidechainorientations
oftheFLMMtetradresiduesineachdomain;theirback- that deviated by <1.0 A˚ in most structures; the smallest
bone structures were closely aligned (Figure 5B). The deviations were observed for M residues. For residues
rmsdfortheCaatomsoftheFLMMtetradineachdomain F19,L32, and F92,deviations ranged from1.2 to 1.4 A˚.
was0.208A˚,andthisvaluewas0.573A˚ foracomparison In these, F19 and F92 were rotated by (cid:4)90(cid:3) relative to
ofthewholedomain.Thus,theFLMMresiduesineachdo- the orientation observed in 2HQW (Figures 6F and 6H),
1606 Structure15,1603–1617,December2007ª2007ElsevierLtdAllrightsreserved
Structure
CalmodulinBoundtoNMDARNR1C1Region
Figure4. DistributionofCaMN-andC-DomainContactsintheCaM-NR1C1pComplex
(A)N-domainresidues%4.5A˚ ofNR1C1pshownassticks;17contactsweremadewithNR1residues875–885(gray),and19contactsweremade
withresidues885–896(black).
(B)SequencemapofCaMresidues%4.5A˚ofNR1C1p.ResiduesinNR1C1pthatmakethehighestnumberofcontactsexclusivelywiththeCdomain
(F880)andtheNdomain(T886)areboxed;theERretentionsignalisunderlined.
(C)C-domainresidues%4.5A˚ofNR1C1pshownassticks;27contactsweremadewithresidues875–885,and7contactsweremadewithresidues
885–896.Ca2+ionsandbindingsites(yellowin[A]and[C])aredesignatedI,II,III,andIV.
ThefigurewasmadewithMacPymol.
Structure15,1603–1617,December2007ª2007ElsevierLtdAllrightsreserved 1607
Structure
CalmodulinBoundtoNMDARNR1C1Region
Figure5. ComparisonofFLMMTetrads
inNandCDomainsofCaM
(A)SequencealignmentoftheCaMNdomain
(1–75) and C domain (76–148). Blue boxes
highlightF19,L32,M51,andM71;redboxes
indicateF92,L105,M124,andM144.Yellow
boxes indicate calcium-binding sites. Resi-
duescontactingK875arepurpleandunder-
lined.
(B)TheCaMNdomain(blue;residues8–73)
andCdomain(red;residues81–146)aligned
accordingtoCaatoms(green)oftheirFLMM
tetrad residues. Ca2+ ions and binding sites
areyellow.
(C)ComparisonofFLMMresiduesidechains
(sticks) after alignment of Ca atoms (green
spheres).
(D)ElectrondensityofFLMM andF880shown
C
atacontourlevelof1.0s.
ThefigurewasmadewithMacPymol.
andLvariedmostattheCgandCdatoms(seeFigure6G; theFLMM tetrad.Astructuralalignment ofthesecom-
C
Table S1, see the Supplemental Data available withthis plexes according to the Ca atoms of the FLMM (Fig-
C
articleonline). ure7B)revealedthattheorientationsoftheresiduecon-
tacting the majority of these FLMM residues in all 13
C
IdentifyingTargetResiduesthatContact structureswerewellconserved.
theFLMMTetradsofCaM Theresidueineachpeptidethatcontactedthehighest
An analysis of the chemical characteristics of the target numberoftheFLMM residuesisboxedinblueinFigures
N
residues that contact FLMM and FLMM revealed that 7Aand8.InthecaseofCaMboundtoCaMKIIa(1CDM),
N C
these tetrads were not used identically by the targets. smMLCK(2BBM),andhRyR1(2BCX),morethanoneres-
Figure7Ashowssequencesofthepeptidein13compact idue contacted an equal number of FLMM residues. In
N
CaM-targetcomplexes.Thesewerealignedaccordingto each structure, the residue that contacted the majority
thepeptideresidue(redbox)thatcontacted thehighest ofFLMM residueswasalsotheonethathadthehighest
N
numberofFLMM residues.In11of13complexes,this number of contacts with all N-domain residues of CaM,
C
residue also made the highest number of contacts with withthe exception of the CaM-CaMKI structure (1MXE),
allC-domainresiduesofCaM(Figure8,redbars).Intwo in which R317 contacted one more residue than M316.
cases, there were 2 residues (Y1627 and F1628 of the However, unlike contacts in the C domain, there were
Ca 1.2 channel in 2BE6, and W3620 and L3623 of otherresiduesinthetargetthathadthesamenumberof
v
hRyR1in2BCX)thateachcontacted3FLMM residues; contacts as these. For example, in the CaM-CaMKIIa
C
F1628in2BE6andW3620in2BCXhadthehighestnum- structure(1CDM),R297,G301,L304,T305,andA309all
berofcontactswithallC-domainresiduesofCaM.In12of contactedthesamenumberofN-domainresidues(4)in
these13structures,theresiduethatcontactedthehighest CaM;however,ofthese,onlyT305andA309contacted
numberofFLMM residueshadalargearomaticmoiety themajorityoftheFLMM residues.
C N
(7 F, 5 W, 1 Y) and two (CaMKII [1CDM] and hRyR1 A large variation was observed in the size, chemical
[2BCX]) had a leucine residue in the cavity defined by characteristics, and spacing of the residues that
1608 Structure15,1603–1617,December2007ª2007ElsevierLtdAllrightsreserved
Structure
CalmodulinBoundtoNMDARNR1C1Region
Figure 6. Statistical Analysis of CaM-
TargetInterfaces
(A–D)Histogramsshowingresiduesinthe(A)C
domain(residues84–146)or(C)Ndomain(res-
idues5–72)ofCaM%4.5A˚fromaboundpep-
tide ordrugin more than 11 of17 compact
CaM-target structures. Bars for residues in
theFLMMtetradsareblack;othersaregray.
Only residues defined in all 17 structures
wereanalyzed.Alignmentof13CaM-peptide
complexesbytheCaatomsoftheir(B)FLMMC
(red)or(D)FLMMN(blue)residues;theirside
chainsareshownassticks.Thedomainsur-
faceof2HQWiscoloredpink(Cdomain)or
lightblue(Ndomain).
(E) A histogram of rmsds for residue side
chainsinFLMM andFLMM in16CaM-target
N C
structurescomparedtothecorrespondingres-
iduein2HQW(seeTableS3).
(F–H)Binsrepresentincrementsof0.2A˚.Com-
parisonofsidechainorientationsofrepresen-
tative FLMM residues with high rmsds from
2HQW(black):(F)F19in2BCX(blue),1SY9(or-
ange), and 1MXE (green); (G) L32 in 2BCX
(blue),2BE6(red),1CKK(orange),and1CDL
(green); and (H) F92 in 1NIW (orange) and
1MXE(green).
ThefigurewasmadewithMacPymol.
contactedthemajorityoftheFLMM residuesrelativeto andmakesthehighestnumberofcontactswiththeNdo-
N
the primary anchor residue at the reference position of mainofCaM,appearstocontactonlytherimofthecavity
‘‘1’’(Yapetal.,2000).In11ofthesequences,theresidue definedbytheFLMM tetrad(Figure7D).
N
contactingFLMM wasahydrophobicaminoacid,butits ToexplorethegeneralavailabilityoftheFLMMtetrads
N
positionvariedfrom10,11,14,16,or17.Thestructuresof tobindingofahydrophobicmoietythatisnotrestricted
CaM-NR1C1p(2HQW),CaM-MARCKS(1IWQ),andCaM- by the orientation and chemical linkage of residues in a
CaMKIIa(1CDM)wereunusualinthattheresiduecontact- target peptide, this analysis of the FLMM tetrads was
ingthemajorityoftheFLMM residueswaspolar(Seror focused to include those in four CaM-drug compact
N
Thr)andatposition7.Astructuralalignmentofthesecom- complexes. Three of these have TFP (Trifluoperazine;
plexes according to the Ca atoms of the FLMM 10-[3-(4-methyl-piperazin-1-yl)-propyl]-2-trifluoromethyl-10-
N
(Figure7C)revealedamuchlargervariationintheposition phenothiazine)boundin3CaM:drugratios(1:1,1:2,1:4),
of the target residue in the FLMM cavity than was ob- andthefourthhasDPD(N-[3,3,-diphenylpropyl]-N0-[1-R-
N
served for the FLMM cavity. In some structures (i.e., (3,4-BIS-butoxylphenyl)-ethyl]-propylenediamine) bound
C
complexes withpeptidesfromMARCKS,CaMKIIa,NR1 in a 1:2 ratio of CaM:drug (1QIV.pdb). In the structures
C1,eNOS,andMyosinVI),theFLMM cavitywasempty witha1:1ratioofCaM:TFP(1CTR.pdb)andwitha1:2ratio
N
or only partially occupied, as illustrated in Figure 7D for (1A29.pdb),onlytheFLMM tetradwasoccupied,andin
C
theCaM-NR1C1pstructure.TheC-domainprimarycon- the structure with a 1:4 ratio of CaM:TFP (1LIN.pdb)
tactresidueofthepeptideinthisstructure(F880)wasob- bothtetradswereoccupiedbyTFP.Astructuralalignment
servedtofilltheFLMM cavityofCaM.However,T886, ofthetwodomainsof1LIN.pdbaccordingtothepositions
C
which contacts the majority (3) of the FLMM residues oftheCaatomsoftheFLMMtetradsillustratedthatTFP
N
Structure15,1603–1617,December2007ª2007ElsevierLtdAllrightsreserved 1609
Structure
CalmodulinBoundtoNMDARNR1C1Region
Figure7. DistributionandOrientationofTargetResiduesContactingCaMinCompactCaM-PeptideComplexes
(A)Sequencesof13peptidesalignedbytheresidue(redbox)thatcontactedthemajorityofFLMMCresidues;thepeptideresiduethatcontactedthe
majorityofFLMMNresiduesisboxedinblue.Thenumbersabovethesequencesdenotethespacingbetweenthese2residues.Tenpeptidesbindto
CaMinanantiparallelorientation;sequencesareshownbyusingthestandardconvention(theN-terminalresidueisleftmost).Threepeptidesnoted
byanasterisk(*)andlistedlastbindtoCaMinaparallelorientation;theirsequencesareshowninreverse.
(BandC)Alignmentof13CaM-peptidecomplexesbytheCaatomsoftheirFLMMresidues.Theseresiduesin2HQWareshownasredspheres;the
sidechainsoftheprimarycontactresidueofthetargetareshownasblacksticks.Thedomainsurfaceof2HQWiscoloredpink(Cdomain)orlightblue
(Ndomain).
(D)FLMMpocketoccupancyin2HQW.FLMM (red)andFLMM (blue)residuesofCaM,aswellasF880(black),T886(black),andF891(gray)of
C N
NR1C1pin2HQWareshownassticks.
(E)DrugoccupancyoftheCaMdomains.AlignmentoftheNdomain(blue,residues8–73)andCdomain(red,residues81–146)ofCaMina1:2
DPD:CaM(1QIV.pdb)bytheCaatomsoftheirFLMMresidues.DPDboundtotheCaMNdomainisblack,andDPDboundtotheCdomainisgreen;
thetransparencyofdomainswas0.5.
ThefigurewasmadewithMacPymol.
wascapableofbindingbothFLMMcavitiesinthesame in the DPD complex versus 0.573 A˚ in the NR1C1p
orientation, and that the two domains of CaM bound to complex).
TFPhavesimilarstructures(rmsdof0.492A˚).Thissimilar-
itybetweendomainswasalsoobservedintheCaM-DPD DISCUSSION
structure,inwhichbothFLMMtetradswereoccupiedwith
thesamemoietyofaDPDmoleculeinthesameorienta- TheC1regionoftheNR1subunitoftheNMDAreceptor
tion(Figure7E).TheNandtheCdomainsofCaMinthis has been shown to regulate receptor trafficking and
structure align nearly as well as the domains of CaM decreasePKC-inducedreceptorpotentiation.Thestruc-
when bound to NR1C1p (Figure 5B) (rmsd of 0.652 A˚ tureof(Ca2+) -CaMboundtoNR1C1p(2HQW)presented
4
1610 Structure15,1603–1617,December2007ª2007ElsevierLtdAllrightsreserved
Structure
CalmodulinBoundtoNMDARNR1C1Region
Figure8. InterfaceAnalysisof13CaM-PeptideComplexes
ResiduesintheNdomain(gray)andCdomain(black)ofCaMwithin4.5A˚ ofapeptideresiduedeterminedwithCSU.Redindicatesthepeptideres-
iduecontactingthehighestnumberofC-domainresidues;blueindicatesthepeptideresiduecontactingthehighestnumberofN-domainresidues.
heredemonstratesthatC1isa1-7motif,anditindicates tation of NR1C1 mutants that would disrupt association
which residues contribute to the interface between with CaM and serve to test the role of CaM in NR1
CaMandNR1C1.Itprovidesafoundationfortheinterpre- trafficking.
Structure15,1603–1617,December2007ª2007ElsevierLtdAllrightsreserved 1611
Structure
CalmodulinBoundtoNMDARNR1C1Region
PhysiologicalSignificanceofthe highlightedagroupof7hydrophobicresidues(4M,2F,
CaM-NR1C1pComplex 1L)ineachdomainofCaMthatsurroundthehydrophobic
NR1subunitsareexpressedinan(cid:4)10-foldexcessover pocketsinthesestructures.Onthebasisofacomprehen-
NR2subunitsinthecell;however,only40%–50%ofthese sive statistical analysis of the contact distances in 17
NR1subunitsreachthecellsurfaceinculturedhippocam- wrap-around CaM-target structures, we have identified
pal neurons (Okabe et al., 1999). There are eight splice theFLMM tetradasprongsthatholdahydrophobicres-
C
variantsofNR1,andthemajorvariantinthebraincontains idueinallcanonicalCaM-targetcomplexesstudiedhere,
theC0-C1-C2regions(MoriandMishina,1995);however, whereasthecorrespondingFLMM tetradisnotaswell
N
thisvarianthasthelowestfractionofcell-surfaceexpres- contacted.
sion(Okabeetal.,1999).ERretentionoftheNR1subunit
ismediatedbyasequenceofthreecontiguousRresidues ConservationofFLMMTetradsinCaMSequences
ontheC1regionandiscontrolledbythephosphorylation GiventheprevalenceoftheFLMMtetradsinthetarget-
ofS896aftertheERretentionsignal(Scottetal.,2001). bindingpocketsofCaM,itwasexpectedthattheseresi-
Thestructureof(Ca2+) -CaMboundtoNR1C1pshowed dueswouldbeconservedinthesequenceofCaMacross
4
thattheERretentionsignalandS896werenotoccluded species.Comparisonof102CaMsequences(Figure9)re-
byCaM.Whilethisstructurealonecannotruleoutthepos- vealedthat4ofthe8FLMMresidues(F19,L32,F92,and
sibility ofadirectroleofCaMinERretention, itismore M124) were completely conserved, and 2 (M51, L105)
likely that CaM might serve as an indirect modulator by were 99.98% conserved: position 51 was M in all but
interacting with a kinase or other protein that has a role two sequences (Calm_Yeast and Calm_KLULA have
inERretention. L), and position 105 was L in all but two sequences
StudiesofNR1splicevariantscontainingtheC1region (Calm2_PethyhasVandCalm_MouschasW;seeTable
showedthatthisregionwasnecessaryandsufficientfor S2forcompleteanalysisandaccessionnumbers).Inthe
the formation of discrete subcellular receptor clusters 102 CaM sequences analyzed, this was the only occur-
thatareassociatedwiththeplasmamembranewhenex- renceofWatanyposition.Itispossiblethatasequencing
pressedinfibroblastcells(Ehlersetal.,1995).Phosphor- ambiguity could account for the two substitutions
ylationofS890withintheC1regionbyPKCdisruptsthe observed at this position. Codons for W (UGG) and V
receptor-enrichedclusters,resultinginanevendistribu- (GUG) differ by a single base from the sequence for L
tionoftheNR1subunit(Tingleyetal.,1997).Thisresidue (UUG),raisingthepossibilitythatL105isalsocompletely
hadonly41%solventaccessibilitywhenincomplexwith conserved.
(Ca2+) -CaM,anditshydroxylgroupwaswithin4A˚ from Thehydrophobicityoftheremaining2FLMMtetradres-
4
thesulfurgroupsofM36andM51ofCaM.Wehaveshown idues(M71,M144)correspondingtothefourthpositionof
that(Ca2+) -CaMbindsNR1C1pwithahighaffinity(K = thetetradinbothdomainsofCaM(Figure5A)washighly
4 D
2nM).Ehlersandcoworkers(Ehlersetal.,1996b)reported conserved among the 102 sequences compared; how-
comparableaffinitiesforNR1C1pandalargerfusionpro- ever,these2residuesshowedhighersequencevariation
teinofNR1C1p((cid:4)150kDa;K =4nM).Together,these than the other 6 FLMM residues. Position 71 was M in
D
datasuggestthatCaMcanprotectthisregionfromother 62sequencesandLintheother40.Position144wasM
proteinsinthepresenceofcalcium. in67sequences,Vin23,Lin10,andIin2.Thesmaller
size of the variant side chains may allow the pocket to
CommonFeaturesofHydrophobicCavities accommodatelargeranchorresidues.
inCaMDomains
Early studiesofCaM-peptide interactions demonstrated ConservationofFLMMTetrads
the importance of hydrophobic residues found on both A search of the SWISS-PROT knowledgebase and the
thetargetandCaM,particularlytheroleoftryptophanin Protein Data Bank (PDB) for all proteins with identical
apeptideandthemethionine‘‘puddles’’oftheCaMhy- spacing of the primary sequence for FLMM (e.g., F-
drophobicclefts(O’NeilandDeGrado,1990).Subsequent (x12)-L-(x18)-M-(x19)-M) with PROSITE (Sigrist et al.,
structural comparisons have concluded that large hy- 2002) on the ExPASy Proteomics Server (http://ca.
drophobic residues of the target bind the hydrophobic expasy.org)(Gasteigeretal.,2003)identified361nonre-
pocketsofCaM,andthatorientationofbindingisdeter- dundant sequences in these databases (i.e., for se-
minedbytheelectrostaticcharacteristicsofthedomains quences found both in the PDB and the SWISS-PROT
ofCaM(cf.Bhattacharyaetal.,2004;HoeflichandIkura, knowledgebase, the PDB sequence was omitted; Table
2002;IshidaandVogel,2006;VetterandLeclerc,2003). S3).Ofthese,132weresequencesofCaMwith2listings
TherolesofindividualMetresiduesonCaMinenzymeac- peraccessionnumber,1foreachdomain.Thereweresix
tivation have been investigated via mutagenesis (Chin structures available in the PDB of non-CaM sequences
etal.,1997).Dynamicssimulationsfurthersupportthethe- that contained an identical FLMM primary sequence:
sisthatasetofconservedmethioninesineachdomainof CaM-like protein 3(1GGZ-A), the C domain of CaM-like
CaMareflexibleandallowforaccommodationofvariable protein 5 (2B1U-A), the N domain of centrin (caltractin;
peptideresidues(Fiorinetal.,2006).Astructuraloverlayof 2AMI-A), cytochrome P450 (2IJ5-A), aspartyl-rTNA syn-
the domains of CaM in seven wrap-around CaM-target thase(1C0A-A),andtheMchainofthephotosyntheticre-
structures available in the PDB (Ishida and Vogel, 2006) action center (1AIJ-M). Alignment of Ca atoms of the
1612 Structure15,1603–1617,December2007ª2007ElsevierLtdAllrightsreserved
Description:Zeynep Akyol Ataman,1 Lokesh Gakhar,1 Brenda R. Sorensen,1 Johannes W. Hell,2 and gous domains (N and C); each domain has two EF-hand.