Table Of ContentDiseaseMarkers30(2011)197–212 197
DOI10.3233/DMA-2011-0775
IOSPress
Associations of single nucleotide
polymorphisms in the adiponectin gene with
adiponectin levels and cardio-metabolic risk
factors in patients with cancer
a a b a
RashaMazenAlKhaldi ,FahdAlMulla ,ShafikaAlAwadhi ,KusumKapila and
a,∗
OlusegunA.Mojiminiyi
aDepartmentofPathology,KuwaitUniversity,Safat,Kuwait
bDepartmentofMedicine,FacultyofMedicine,KuwaitUniversity,Safat,Kuwait
Abstract.Objectives: Theaimsofthisstudyareto(1)studytheinfluenceofpolymorphismsinadiponectingeneonadiponectin
levelsandpotentialassociationswithbreast,prostateandcoloncancer;(2)investigatetheassociationsofadiponectinlevelswith
otheradipokinesandbreast,prostateandcoloncancers.
Subjects: Wemeasuredfastingadiponectin,leptin,insulin,Sexsteroidsin132(66females,66males)cancerpatientsand68age
andsexmatchedapparentlyhealthysubjects. BodyMassIndex(BMI)andwaistcircumferencewereusedasindicesofobesity.
InsulinResistancewasassessedusingHomeostasisModelAssessment(HOMA).Threesinglenucleotidepolymorphisms(SNP
rs182052 (G-10066-A), SNPrs1501299 (276G >T),SNPrs224176 (45T>G)inadiponectin genewerestudied usingReal
TimePolymeraseChainReaction.
Results: GGgenotypeofSNPrs1501299wassignificantlyassociatedwithhigherlevelsofadiponectin(OR=1.2,95%CI(1.03–
1.3), p = 0.02); breast (OR= 8.6, 95%CI(1.03–71), p = 0.04), colon cancers (OR= 12, 95%CI(1.2–115), p = 0.03). GT
genotypewasalsoassociatedsignificantlywithcoloncancer(OR=2.6,95%CI(1.1–6),p=0.03). HoweverSNPrs224176was
associatedwithonlybreastcancer.
Conclusion: OurresultsdemonstratethatadiponectingeneSNPrs1501299andSNPrs224176maybethepredisposingfactorsin
somecancersbutourresultsdifferfromwhathasbeenreportedinotherpopulationssuggestingacomplexrelationshipbetween
geneticvariationsandphenotypicadiponectinlevels.
Keywords:Adiponectingene,SNPs,cancer,insulinresistance,adipokines
ListofAbbreviations SHBG SexHormoneBindingGlobulin
HOMA Homeostasismodelassessment
DNA Deoxyribonucleicacid
RNA Ribonucleicacid
SNPs SingleNucleotidePolymorphisms AD AllelicDiscrimination
KCCC KuwaitCancerControlCenter CI ConfidenceInterval
BMI BodyMassIndex OR OddsRatio
ELISA EnzymeLinkedImmunosorbentAssay ANOVA One-wayanalysisofvariance
FSH FollicleStimulatingHormone SPSS StatisticalPackageforSocialSciences
LH Luteinizinghormone mRNA messengerribosenucleicacid
1. Introduction
∗Corresponding author: Dr. O.A. Mojiminiyi, Department of There is emergingevidence that adipokinesare as-
Pathology, FacultyofMedicineKuwaitUniversity, POBox24923
sociatedwithseveraltypesofobesityrelatedcancers.
Safat,Kuwait,Code13110.Tel.:+96525319476;E-mail:segunade
@yahoo.com. Adiponectin is an adipokine that is involved in the
ISSN0278-0240/11/$27.502011–IOSPressandtheauthors. Allrightsreserved
198 R.M.AlKhaldietal./AssociationsofadiponectingeneSNPsincancer
control of glucose, fat metabolism and insulin sen- resistance. Data of association analysis of these two
sitivity with anti-diabetic, anti-atherogenic and anti- previouslymentionedSNPs havebeeninconsistentin
inflammatory activities [1]. In addition to its func- differentpopulations. Forexample,astudyinGerman
tionasaninsulinsensitizinghormone,adiponectinmay population has shown significant association of 45T
playaroleintheregulationofcellgrowthanddeath[2, > G variant with obesity and insulin sensitivity [15]
3]. It inhibits proliferation of cell through selective- whereas a Japanese study found no such association
lybindingandsequesteringthegrowthfactorssuchas withthesameSNP[13].
PlateletDerivedGrowthFactor(PDGF),Basicfibrob- Ontheotherhand,Haraetal.,observedanassocia-
last growth factor and other factors, thus suppressing tionbetweenSNPsatpositions45and276andType2
their interaction with membrane receptors. Further- diabetesintheJapanesepopulation[12]. Haraandcol-
more, adiponectin has been suggested to inhibit the leagues reported that subjects with the G/G genotype
activationofnuclearfactorkappa-light-chain-enhancer atposition45ortheG/Ggenotype,atposition276had
ofactivatedBcells(NF-κB),atranscriptionfactorthat a significantly increased risk of type 2 diabetes com-
isinvolvedininitiatingsurvivalsignalingpathway[3]. paredwiththosehavingtheT/Tgenotypeatpositions
Adiponectin also promotes apoptosis through activa- 45 and 276, respectively, indeed more than 40% of
tion of a cascade of caspases −8, −9 and −3, which JapaneseindividualshavethehighriskG/Ggenotype
leads to cell death. Hence, it has been proposed that which makes subjects genetically prone to decreased
adiponectinisananti-canceradipokinethathasprotec- adiponectinlevels[12].
tiverolesagainstcarcinogenesis. Inanotherstudy,GalleleofSNPrs224176andG/G
Lowlevelsofadiponectinhavebeenfoundinanum- genotypeofSNPrs1501299wereassociatedwithim-
berofobesityrelatedcancersaswellascancersasso- paired glucose tolerance in Spanish population [16].
ciatedwithinsulinresistance[4]suchasbreast[5–7], The study has also reported that the G/G genotype
prostate [8] and colon [9]. It has been suggestedthat for SNP rs1501299 was associated with lower serum
reducedlevelsofadiponectinleadtothedevelopment adiponectinlevelscomparedtoG/TandT/Tgenotype
ofinsulinresistanceandcompensatorychronichyper- andhence suggestingthatSNP rs1501299may affect
inuslinemia which in turn lead to reduced liver syn- theexpressionofadiponectin[16].Reportsontheasso-
thesis of IGFBP1 and IGFBP2. As a result levels of ciationsoftheseSNPshavealsobeeninconsistentand,
bioavailableIGF1increasesfavoringthedevelopment inagreementwithreportsfromJapanese[13]andGer-
oftumors. Inparallel,chronichyperinuslinemiadown man[17]populations,theGalleleofSNPrs224176did
regulates the hepatic synthesis of SHBG, thereby in- notresultinanychangeinadiponectinconcentrations
creasingfreesexhormones. Inaddition,lowlevelsof intheSpanishpopulation[15].
adiponectin may probably up-regulate fatty acid syn- Astheassociationsofthesecommonlystudiedpoly-
thase(FAS) a keylipogenicenzymethathasbeenas- morphisms in the adiponectin gene with the risk of
sociated with breast, prostate and colon cancers [10]. obesity related cancershave not been established and
A novel signaling pathway linking adiponectin with publisheddataonotherassociationshavebeenincon-
carcinogenesis is JNK pathway. This belongs to the sistent, the aims of this study are to (1) investigate
mammalian mitogen activated protein (MAP) kinase theassociationbetweenadiponectinconcentrationand
family that acts as a stimulator for different survival other adipokines and breast, prostate and colon can-
signalingpathwaysincludingtumordevelopment[11]. cers, (2) study the influence of polymorphismsin the
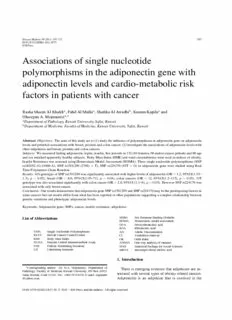
Figure 1, summarizesthe mechanismsthroughwhich adiponectin gene on adiponectin levels and potential
obesityrelatedcancerscouldbemediated. associationswithbreast,prostateandcoloncancers.
Screeningoftheadiponectinencodinggenehasled
to the discovery of genetic variants and single nu-
cleotide polymorphisms(SNPs) that have been found 2. Materialsandmethods
to be associated with reduced circulating levels of
adiponectin, Type 2 diabetes, insulin resistance and 2.1. Patientsandcontrols
obesity[12–14]. TwocommonSNPshavebeenstud-
ied extensively in the literature the substitution of T As part of the protocol approved by local ethical
toGinexon245T>G (rs2241766)andsubstitution committeeinaccordancewiththeethicalstandardsin
of G to T in intron 2 276G > T (rs1501299) along Helsinki declaration, we obtained fasting blood sam-
withtheirassociationwithdiabetes,obesityandinsulin ples from 132 (66 females, 66 males) cancer patients
R.M.AlKhaldietal./AssociationsofadiponectingeneSNPsincancer 199
Fig.1.Mechanismsthroughwhichobesityrelatedcancercouldbemediated.
whowererecruitedfromKuwaitCancerControlCenter cumference(cm)atthefemoralgreatertrochanterand
(KCCC).Thediagnosisofcancerwasmadeonclinical themaximalglutealprotuberancewerealsotaken. All
basis, radiological findings and histological findings. anthropometric measurements were taken at the time
Patientswererecruitedimmediatelyafterdiagnosisand ofsubject’senrollmentandblooddraw.
beforestartingtreatment. Sixtyeight(34femalesand
34males)ageandsexmatchedapparentlyhealthyvol- 2.3. Laboratoryanalysis
unteerswereenrolledfromKuwaitBloodBankCenter.
Exclusioncriteriaincludedrefusaltoparticipateinthe Plasma adiponectin concentrations were measured
studyaswellasthepresenceofotherendocrinedisease byEnzymeLinkedImmunosorbentAssay(ELISA)Kit
(Diabetes, heart failure, renal failure) or ingestion of (HumanAdiponectinELISAKit,LincoResearch,Mis-
anymedication(suchasthiazolidinedione)thataffects souri,USA).Theassaysensitivityis2ng/mL.Intra-and
adiponectin,hormonesorlipidprofileatthetimeofthe Interassaycoefficientsofvariationwere4%and4.1%
study. Patients with previoushistory of othercancers respectivelyatadiponectinconcentrationof7μg/mL.
and recent history (within the previous 1–6 months) Plasma leptin was measured by using the (DSL-10-
of weight gain or loss of 5% or more of their current 23100ACTIVEHumanLeptinELISAkit). Theassay
weightwerealsoexcludedfromthestudy. sensitivity is 0.05 ng/mL, whereas the intra and inter
assay coefficients of variation were 3% and 3.3% re-
2.2. Anthropometricmeasurements spectively at leptin concentration of 23 ng/mL. Lev-
elsofFollicularStimulatingHormone(FSH),Luteiniz-
Body weight (Kg) was measured in light clothing ing Hormone (LH), Total Testosterone and Sex Hor-
without shoes and height (cm) was measured with a mone Binding Globulin (SHBG) were determined by
stadiometer. Subsequently body mass index (BMI) usingIMMULITE1000Kits(SiemensHealthcareDi-
(Kg/m2)wascalculatedaccordingtothefollowingfor- agnostics, Deerfield, USA). Fasting plasma glucose
mula: BMI=Weight(Kg)/Height(m2). Waistcircum- was analyzed on an automated analyzer (Beckman
ference(cm)attheleveloftheumbilicusandhipcir- DXC, Beckman Corporation, Brea, CA, USA) and
200 R.M.AlKhaldietal./AssociationsofadiponectingeneSNPsincancer
Table1
Demographic,anthropometricandbiochemicalcharacteristicsofthestudypopulation
Parameter Cancerpatients(n=132) Controls(n=68) p-Value
TypeofCancer Breast(n=60) Prostate(n=14) Colon(n=58)
Age(years) 49±2.4 59±8.7 53±7.7 60±5.2 NS
BMI(Kg/m2) 28.8±4.1 27.7±3 27.9±6 26.0±3 NS
WC(cm) 102±15 108±14 106±14 102.5±9 NS
Systolicbloodpressure(mmHg) 131±20 131±20 131±20 130±12 NS
Diastolicbloodpressure(mmHg) 80.6±6 80.6±6 80.6±6 81±8 NS
Plasmaglucose(mmol/L) 6.8±6 6.8±2 7.3±1.7 5.8±0.5 <0.0001
TotalCholesterol(mmol/L) 6±1 7.1±2 5.6±1 6.6±1.1 NS
HDL(mmol/L) 1.2±0.5 1.4±0.9 1.3±0.4 1.2±0.07 NS
LDL(mmol/L) 3.9±2 4.9±1 3.7±1 4.4±0.6 NS
Adiponectin(µg/mL) 8.45±4 10.4±3 8.6±4 4.1±2 <0.0001
Insulin(µIU/mL) 13±4.8 10.73±6 10.4±8.6 11.0±1.5 <0.0001
HOMA 4.3±0.4 3.4±0.5 3.3±0.2 3.0±0.7 <0.0001
BMI=BodyMassIndex,WC=WaistCircumference,HOMA=HomeostasisModelAssessment.
fasting serum insulin was determined by an enzyme- izedbylogtransformationtouseparametrictests. De-
linked immunosorbent assay (ELISA) (DSL-10-1600 scriptive statistics namely mean ± SD and 95% con-
ACTIVE, Diagnostics Systems Laboratories, Texas, fidence interval(CI) were used as appropriate. Com-
USA).HomeostasisModelAssessment(HOMA)was parison between mean values in the 2 groups (Pa-
usedasanassessmentforinsulinresistancebyusingthe tients and Controls) was evaluated using student’s t
HOMA-IR=glucose(mmol/L)Xinsulin(μIU/mL)/by test. Spearmancorrelationcoefficientwasusedtode-
22.5. scribe the associations between continuous variables
of interest; the partial correlation analyses were used
2.4. DNAisolation to control for the effects of age, sex and BMI. The
association between adiponectin genotypes and can-
NucleospinBloodKit(Macherey-Nagel,Neumann- cer risk was determined by calculating the odds ratio
Neader, Germany) was used to extract Deoxyribonu- (OR) and 95% confidence interval (CI) using multi-
cleic acid (DNA) from citrated bloodsamples, which variateand binarylogistic regressionwith adjustment
providedan expectedyield range of 4–6 μg of DNA. fortheconfoundingeffectsofage, sexand BMI. The
OnceDNAwasisolated,theDNAquantityandquality relationship between adiponectin, genotypesand bio-
werecheckedusingaspectrophotometer. chemical findings were evaluated by one-way anal-
ysis of variance (ANOVA). All the statistical analy-
2.5. Genotypingofpolymorphismsinadiponectin ses were performed with the Statistical Package for
gene Social Sciences (SPSS Inc. Chicago, USA), version
16.0. Thecriterionforstatisticalsignificancewasp<
AllelicDiscrimination(AD)ofadiponectingenein 0.05. Hardy-WeinbergEquilibriumwasusedtoevalu-
200 samples was done by using 7500 fast Real Time ate the allele frequenciesusing the followingfacility:
PolymeraseChainReaction(AppliedBiosystems,Fos- www.oege.org/software[18]. The powerof the study
ter City, California, USA). SNPs were chosen based wascalculatedusingthefollowingfacility: http://hed-
on the most commonly reported in the literature and wig.mgh.harvard.edu/samplesize/size.html[19]. The
using Applied Biosystems SNP browser version 3.5. SPSSsoftwarewasalsousedtorecodeforeachcom-
Three SNPs were studied as follows: SNP G-10066- bination.
A (rs182052) in intron 1, SNP 276G>T (rs1501299)
in intron 2, SNP 45T>G (rs224176) in exon 2. The
selection of the SNPs was based on previous studies 3. Results
showingassociationwithcancerinotherpopulation.
3.1. Demographic,anthropometricandbiochemical
2.6. Statisticalanalysis characteristicsofthestudypopulation
Data were tested for normality using Kolmogorov Table1summarizesthedemographic,anthropomet-
Smirnovtest. Non-parametricvariableswerenormal- ricandbiochemicalcharacteristicsofthestudypopula-
202 R.M.AlKhaldietal./AssociationsofadiponectingeneSNPsincancer
Fig.2. Differences inadiponectin concentration according toHOMAindex. Thebarsrepresentthe95%confidenceinterval ofadiponectin
concentrationandthecirclewithinthebarsrepresentsthemean.
tion. Atotalof132(66femalesand66males)patients cer patients had significantly higher (p < 0.0001)
with breast, prostate and colon cancer and 68 (34 fe- insulin levels and significantly higher (p < 0.0001)
malesand34males)apparentlyhealthysubjectswere HOMAindexasshowninTable2.
includedinthisstudy.
3.3.1. Stratificationofthestudypopulationaccording
3.2. Adiponectin tothesex
Table 2 demonstrates the differences between obe-
Table 2 demonstrates a significant difference in sity related parameters in control and patient groups
levels of adiponectin in control and patient groups. of women and men respectively. We found a signifi-
Cancer patients had significantly (p < 0.0001) high- cantdifferencein levelsofadiponectinin controland
er adiponectinconcentrationsthan apparentlyhealthy patientgroupsoffemalesandmalesrespectively. Fe-
subjects. Based on BMI and HOMA-sub grouping, malecancerpatientshadsignificantly(p=0.03)higher
levelsofadiponectinwerelowerinobesethaninnon- adiponectinconcentrationsthandoapparentlyhealthy
obesesubjects(6.9±4.1μg/mLvs.7.9μg/mL)(p = subjects. Inmalecancerpatientslevelsofadiponectin
0.03). Inaddition,insulinsensitivesubjects(HOMA< were significantly higher (p < 0.0001)than in appar-
2) had higheradiponectinlevelsthan insulin resistant entlyhealthysubjects.
subjects (HOMA > 2) (Fig. 2). Adiponectinshowed
sexualdimorphisminapparentlyhealthysubjects,with 3.3.2. Insulinandinsulinresistance
female subjectshaving higheradiponectinlevelsthan Significant differences in insulin and HOMA were
malesubjects(p < 0.0001)(Fig.3). However,Fig.3 foundbetween patientand controlgroupsof females.
demonstratesthatthissexualdimorphismdisappeared Female cancer patients had significantly higher (p <
incancerpatients(p>0.05). 0.0001) insulin levels and significantly higher (p =
0.008) HOMA index as shown in Table 2. Further-
3.3. Insulinandinsulinresistance more,therewasasignificantdifference(p<0.0001)in
insulinandHOMAbetweencontrolandpatientgroups
Significant differences in insulin and HOMA were of males. Males with cancer had significantly higher
foundbetweenpatientandcontrolgroups;indeedcan- (p < 0.0001) serum insulin levels and subsequently
R.M.AlKhaldietal./AssociationsofadiponectingeneSNPsincancer 203
Fig.3.SexualDimorphismofadiponectininthestudysubjects. Thebarsrepresentthe95%confidenceintervalofadiponectinconcentrationand
thecirclewithinthebarsrepresentsthemean.
significantlyhigherHOMAindexthandocontrolmale ly with estradiol but with BMI, waist circumference,
subjects(Table2). insulin, HOMA and total testosterone. Total testos-
teronecorrelatedpositivelywithwaist circumference,
3.3.3. Hormonaldifferencesbetweenpatientsand insulin levels and HOMA indexwhereasit correlated
controls negativelywithadiponectinandestradiol.
Femalecancerpatientshadsignificantlyhigher(p< Afteradjustmentforage,sexandBMI,adiponectin
0.0001)testosteronelevelsthandoapparentlyhealthy correlatedpositivelywithestradiolandnegativelywith
female subjects as shown in Table 2. On the other insulinlevels,HOMAindexandtestosterone(Table4)
hand,malecancerpatientshadsignificantlyhigher(p< indicating that the correlations between adiponectin
0.0001)estradiol levelsthan do male controlsubjects and these variables are independent of age, sex and
asappearsinTable2. However,malecontrolsubjects BMI.
hadsignificantlyhigher(p<0.0001)testosteronelev-
els than do male cancer patientsas shown in Table 2. 3.3.5. Prevalenceratesofadiponectingenotypesin
AsshowninTable2,levelsofFSHandLHinfemale thestudypopulation
cancerpatientsweresignificantlyhigher(p<0.0001) Table5summarizestheprevalenceratesofadiponec-
than female control subjects. Similar trend was ob- tingenotypeswhichwereinHardy-Weinbergequilib-
served in male subgroups of patients and controls as riumamongthestudypopulation.
shownin Table 2. Althoughpatientsgroups(females
and males) have higher SHBG levels than apparently 3.3.6. Anthropometricandbiochemical
healthysubjects,thedifferenceswerenotsignificantas characteristicsaccordingtoadiponectin
showninTable2. genotypes
Although no significant difference (p > 0.05) was
3.3.4. Correlationbetweenvariables observedinBMIamongadiponectingenotypesofSNP
Table3presentstheresultsofSpearmanrankcorre- rs182052, homozygouscarriers of G/G allele had the
lationsbetweenthevariablesofinterestinallsubjects highestBMI(30±7.4Kg/m2)followedbyA/Ageno-
(patientsandcontrols).InsulinlevelsandHOMAindex types (29.8 ± 5 Kg/m2) and A/G genotypes (25.6 ±
correlated positively and significantly with BMI and 5.8 Kg/m2). In contrast, the highest waist circumfer-
Waistcircumference. Adiponectincorrelatedpositive- encewasfoundinhomozygouscarriersofA/A geno-
204 R.M.AlKhaldietal./AssociationsofadiponectingeneSNPsincancer
Table3
Spearman’srankcorrelationsofvariablesofinterestamongallsubjects
AGE BMI WC INS HOMA ADIPO E2 TES
AGE r 1
BMI r 0.05 1
WC r 0.2∗∗∗ 0.5∗∗∗∗ 1
INS r 0.06 0.3∗∗∗∗ 0.3∗∗∗∗ 1
HOMA r 0.07 0.3 0.3 0.9∗∗∗∗ 1
ADIPO r −0.1∗ −0.1∗ −0.2∗∗ −0.2∗∗∗∗ −0.2∗∗∗∗ 1
E2 r 0.08 −0.07 0.02 −0.2 −0.2 0.2 1
TES r 0.09 −0.03 0.1 0.2∗∗∗∗ 0.1∗∗ −0.2∗∗ −0.2∗∗ 1
∗p<0.05,∗∗p<0.01,∗∗∗p<0.001,∗∗∗∗p<0.0001,No∗=NoSignificance.
Age= years, BMI= Body Mass Index (Kg/m2), WC= Waist Circumference (cm), INS = Insulin
(µIU/mL),HOMA=HomeostasisModelAssessment,ADIPO=Adiponectin(µg/mL),E2=Estradiol
(pg/mL),TES=Testosterone(ng/dL).
Table4
Partialcorrelationcoefficientsamongsubjectsaftercorrectionforage,sex
andBMI
WC INS HOMA ADIPO E2 TES
WC r 1
INS r 0.2 1
HOMA r 0.2 0.9∗∗∗∗ 1
ADIPO r 0.08 −0.3∗ −0.3∗ 1
E2 r 0.08 −0.1 −0.08 0.3∗∗∗∗ 1
TES r 0.04 0.03 −0.01 −0.4∗∗∗∗ −0.2∗ 1
∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, No ∗ = No
Significance.
Age=years, BMI=BodyMassIndex (Kg/m2), WC=WaistCircum-
ference(cm),INS=Insulin(µIU/mL),HOMA=HomeostasisModelAs-
sessment,ADIPO=Adiponectin(µg/mL),E2=Estradiol(pg/mL),TES
=Testosterone(ng/dL).
types(104.2±14.2cm)ofSNPrs182052,followedby Homozygous carriers of the G allele of SNP
G/G (103.1± 16.2cm) and A/G genotypes(101.9± rs182052tendedtohaveloweradiponectinlevelsthan
12.6cm). However,thedifferencesdidnotreachsta- carriers of (A/A) or (A/G) genotypes [(G/G: 4.7 ±
tistical significance (p > 0.05). There was also no 2μg/mL,A/A:5.5±3μg/mL,A/G:6.9±3.7μg/mL)]
significantdifferenceinBMIandwaistcircumference but this differences were not significant. Among the
betweentheSNPrs1501299andSNPrs2241766geno- genotypes of SNP rs2241766, homozygous carriers
types indicating that variations in anthropometric in- of the G alleles had the highest levels of adiponectin
dices are not related to genetic polymorphismsin the comparedwiththeothergenotypesasfollows: [(G/G:
adiponectingene. 7.7±1.8μg/mL,T/T:6.8±4.1μg/mL,G/T:6.6±
4.1μg/mL)].Althoughvariationsininsulin,leptinand
othervariableswereobservedamonggenotypesofSNP
3.3.7. Variationofadiponectinandotherobesity
rs182052andSNPrs2241766,thedifferenceswerenot
relatedparameterswithadiponectingenotypes
statisticallysignificant,hence,datanotshown.
inbothpatientsandcontrols
Thereweresignificantdifferencesinadiponectinlev- 3.3.8. Adiponectingenotypesandmultivariatelinear
els in SNP rs1501299 genotypes (Table 6). Table 6 regressionresults:
showsthathomozygouscarriersof T allele (T/T)had Using multivariatelogistic regressionanalysiswith
the lowest concentration of adiponectin compared to inclusionofageandsexasconfounders,wefoundthat
theG/GorT/Ggenotypes. InsulinlevelsandHOMA (GG) genotype of SNP rs1501299 was significantly
indexalsovariedsignificantlyamongSNPrs1501299 associated with levels of adiponectin (B = 0.2, p =
genotypesasshowninTable6. Althoughleptinlevels 0.013)asshowninTable7. Alsohomozygouscarriers
variedamonggenotypesofSNPrs1501299,thediffer- ofG allelearemorelikelytohavehigheradiponectin
enceswerenotsignificant. levelsthan(T/T)or(T/G)genotypesofSNPrs1501299
R.M.AlKhaldietal./AssociationsofadiponectingeneSNPsincancer 205
Table5
Thedistributionofadiponectingenotypeswithinthestudypopulation
Adiponectingenotype Numberofcontrols(n=68) Numberofpatients(n=132)
SNPrs182052
AA 4(5.9%) 8(6.1%)
AG 52(76.5%) 101(76.5%)
GG 12(17.6%) 23(17.4%)
SNPrs1501299
TT 46(67.6%) 63(47.73%)
GT 21(30.9%) 55(41.67%)
GG∗ 1(1.5%) 14(10.61%)
SNPrs2241766
TT 50(73.5%) 84(63.64%)
TG 18(26.5%) 43(32.58%)
GG None 5(3.79%)
TOTAL 68(100%) 132(100%)
∗SignificantDifferenceinthedistributionofGGgenotypebetweencontrolsandpatients.
(OR = 1.2, 95%CI (1.0–1.3), p = 0.02). We also been evidence that the link between obesity and risk
foundthatthisgenotypeisassociatedwithlowerinsulin of cancer is due, in part, to obesity-associated reduc-
levels (OR = 0.8, 95%CI (0.8–0.9), p = 0.03). But tionincirculatingadiponectinlevels[23]. Tothebest
thisgenotypewasnotfoundtobeassociatedwithany ofourknowledgemostofthestudieshaveshownthat
parameters of obesity (BMI or waist circumference). cancerpatientshadloweradiponectinlevelscompared
SNPrs2241766andSNPrs182052genotypeswerenot to their apparently healthy control subjects. Mant-
foundtobeassociatedwithindicesofobesity,insulin zoresandcolleagues,haveshownthatlowercirculating
resistanceandadiponectin(Table7). Usingbinarylo- adiponectin levels are associated with increased risk
gisticregressionanalysiswithinclusionofageandsex ofbreastcancer[5,6]Furthermore,adiponectinlevels
as confounders, SNP rs1501299was significantly as- havebeendemonstratedtobelowerinprostatecancer
sociatedwithcancerinallpatients(OR=2.195%CI patients compared to benign prostatic hyperplasia or
(1.3–3.6),p=0.004)asshowninTable8. Specifically healthycontrolsubjectsandinverselyassociatedwith
GGgenotypewasstronglyassociatedwithcancerde- histologicalgradeandGleasonscore[24]. Ontheother
velopment(OR=9.3,95%CI(1.2–73,p=0.04)inall hand,Biallargeonetal.,foundnoassociationbetween
patients. Itisalsoassociatedwiththedevelopmentof adipokines,adiponectininparticularandprostatecan-
breastcancer(OR=8.5,95%CI(1.03–71),p =0.04) cer [25]. One prospective nested case control study
andcoloncancer(OR=12.0,95%CI(1.2–115),p = reportedthatplasmaadiponectinlevelswereinversely
0.03). SNPrs2241766wasassociatedwithbreastcan- associatedwiththeriskofcolorectalcancerinmen[9].
cer(OR=2.1,95%CI(1.1–4.1),p=0.03)butnotwith Findingsinthisstudycameasasurprisebecausewe
prostateandcoloncancers. Wehavecorrectedforcon- were expecting to find lower adiponectin in associa-
founderssuchasage, BMIandgender,aswealso in- tionwithobesityrelatedcancersasreportedinstudies
cludedadiponectinasaconfounder,resultsdidnotdif- from other populationsmentioned earlier. Therefore,
ferindicatingthattheassociationbetweenadiponectin ourfindingsof significantlyhigheradiponectinin pa-
SNPswithobesityrelatedcancersisindependentofage tientswithcancerareatvariancewithotherpublished
andsex. However,Table8showsthattheassociation studies. Ourresultscouldbeexplainedbyanumberof
between GG genotype of SNP rs1501299 and breast factors. Firstofallitispertinenttonotethattheasso-
cancer is dependenton age, adiponectin levels, BMI, ciationsofadiponectinwithobesityandobesityrelated
andsex. factorssuchasinsulinresistanceareinagreementwith
what is expected from published studies. For exam-
ple,adiponectinwasinverselycorrelatedwithmarkers
4. Discussion of obesity BMI and waist circumferenceas shown in
Tables3 and 4. Adiponectinalso showed the expect-
Severalpublishedstudieshaveshownanassociation ed significant inverse relationships with insulin resis-
betweenobesityandriskofdevelopingprostate,breast tance and insulin levels as shown in Tables 3 and 4.
and colon cancers [20–22]. Additionally, there has BasedonHOMA sub-grouping,insulinsensitivesub-
R.M.AlKhaldietal./AssociationsofadiponectingeneSNPsincancer 209
jects(HOMA<2)hadhigheradiponectinlevelsthan significantlyassociatedwithindicesofobesityinsharp
insulin resistant subjects (HOMA > 2) as shown in contrasttoreportsfromotherpopulations. Astudyin
Fig.2. HispanicfamiliesreportedthatSNPrs182052wasas-
The higher levels of adiponectin in cancer patient sociatedwithfourofsixobesitymeasurements(BMI,
groupscouldbeduetoslightlylower(butsignificant) Waist, WHR, subcutaneous fat) [17] and Yang et al.,
waist circumference in the patient group particularly (2007)reportedthatGalleleofSNPrs1501299wasas-
in male cancer patients as shown in Table 2. Circu- sociatedwithreducedriskofobesity[32]. Inaddition,
lating adiponectinin this study has been shown to be SNPrs2241766,eitherindependently[31]orasahap-
negativelycorrelatedmoresignificantlywithwaistcir- lotypetogetherwithSNPrs1501299[15],wasstrongly
cumferencethanBMIinagreementwithotherprevious associated with obesity and insulin resistance in non-
studies[26,27]. Asestradiolcorrelatedpositivelywith diabetic German and Italian Caucasians. However,
adiponectin,anotherimportantfactorthatcouldexplain thereareinconsistentresultsfromvariouspopulations
thehighlevelsofadiponectininmalecancerpatientsis astheseassociationswerenotfoundinFrench[14]and
thesignificantlylowertestosteroneandrelativelyhigh- Swedish [34] populations suggesting high variability
erestradiollevelswhichinturncouldresultinincreased inphenotypicexpressionamongdifferentpopulations.
productionof adiponectinin male cancer patients. In Thelackofsignificantassociationbetweenthestudied
relationtosexsteroids,thefindingsinfemalesarediffi- SNPsandindicesofobesityisreflectedintheirlackof
culttoexplain,femalecancerpatientshadsignificantly significantassociationswithleptin(Table6).
higher testosterone levels which is expected to result Inthisstudy,thedifferencesincirculatingadiponec-
in loweradiponectinlevelsdueto the factthattestos- tinbetweengenotypesofSNPrs182052werenotsta-
teronesuppressesadiponectinproductionandalsolim- tisticallysignificant. Thismaybeexplainedbythefact
its the production of HWM adiponectin in vivo and thatSNPrs182052isanintronicSNPthatmaynotaf-
inculturedadipocytes[28–30]. However,thereis the fectthe proteinproductof adiponectingene. Howev-
possibilitythattheeffectoftestosteroneonadiponectin er, we havealso foundthatthe G/G genotypeofSNP
is differentin males and females. More significantly, rs1501299 resulted in higher serum adiponectin lev-
females with cancer had higher estradiol due to their els. Our findings are in contrast with results report-
higherBMIwhichcouldresultinhigherrateofperiph- ed in Japanese and Spanish populations that showed
eralaromatization. Thus,itishighlyprobablethatthe lower adiponectin levels in G allele carriers of SNP
effectofestradiolonincreasingcirculatingadiponectin rs1501299[13,14]. The mechanism by which the in-
overrides a tendency of higher testosterone to reduce tronicSNPrs1501299canaffecttheexpressionlevelof
adiponectin. Obviously,sexsteroidsplayaroleinreg- thegeneandincreasecirculatingadiponectincouldbe
ulation of adiponectin levels and the regulation could explainedbyseveralfactors. Itmightbeduetoaspecif-
differinmenandwomen. iclinkagestructureoritmayactwithcertainenviron-
Discrepanciesbetweenourresultsandpreviousstud- mentalfactors and ultimately affectthe expression of
iescouldalsobeexplainedbyethnic/racialdifferences; thegene(gene-environmentalinteraction). Onestudy
indeedpreviousstudieswereconductedonpopulations has indicated that SNP rs1501299 is in moderate to
fromUSA,EuropeandJapan. Geneticstudieshaveas- strong Linkage Disequilibrium with several polymor-
sociatedseveraladiponectingenepolymorphismswith phismssuchasinsertion/deletion+2019placedinthe3
adiponectin levels, obesity, insulin resistance, Type un-translatedregion(UTR)–aregionwhichhasbeen
2diabetes,hypertensionandcardiovasculardiseasesin showntoplayaroleinthecontrolofgeneexpression
somebutnotallstudies[12–14]. Inthisstudywehave by binding proteins that regulate mRNA processing,
found that SNP 276G > T was significantly associ- translationordegradation[33]. Ontheotherhand,we
ated with cancer in all patients while SNP rs2241766 showedthatneitherTnorGallelesofSNPrs2241766
wasassociatedwithbreastcancerbutnotwithprostate resulted in any change in adiponectin concentrations.
andcoloncancers. Furthermore,incontrastwithwhat As SNP rs2241766 is a silent substitution (GGT →
hasbeenreportedinotherpopulations,the(GG)geno- GGG),itmayindicatethatthisvariantdoesnotaffect
type of rs1501299 was significantly associated with the expression of the gene. Our findings agree with
higher levels of adiponectin and rs2241766 and SNP reportsfromJapaneseandGermanpopulations[11,28]
rs182052 genotypes were not found to be associated which found no association between SNP rs2241766
with adiponectin,indicesof obesity andinsulin resis- and adiponectin but at variance with Menzaghiet al.,
tance. All the SNPs evaluated in this study were not whoshowedassociationofinsulinresistancewiththe
210 R.M.AlKhaldietal./AssociationsofadiponectingeneSNPsincancer
Tallelewithdecreasedserumadiponectinlevels[37]. firmedbyotherstudies,therecouldbeaparadigmshift
The conflicting association results in various popula- in the belief that adiponectin SNPs that cause higher
tionssuggestacomplexrelationshipbetweenvariations adiponectinlevelsareassociatedwithdecreasedriskof
in the adiponectin gene and phenotypic adiponectin cancer.
levels. Thiscouldexplainthe differencebetweenour There are some limitations to this study. Although
studyandthatofKaklamanietal.,whofoundincreased thenumberofstudysubjectsgivesthestudyastatisti-
breast cancer risk in adiponectin gene SNPS that are calpowerof90%todetectthechangesinadiponectin
associatedwithloweradiponectinlevelsanddecreased levels,thenumberofsubjectswithcertaingenotypesis
riskofcancerwithSNPsthatincreaseadiponectinlev- smallandstudiesinlargerpopulationsarerequiredto
els [37]. In this study we found a significant asso- confirmourfindings. Theassayusedinthisstudyiden-
ciation between SNP rs1501299and cancer in all pa- tifiestotaladiponectinandcannotdistinguishbetween
tients, breast, and colon cancer as shown in Table 8. the adiponectin isoforms, high- and low-molecular-
Our finding of an association between an adiponetin weight complexes which may have different biolog-
gene SNP that causes higher adiponectin and cancer ical activities. Studies have also shown that post-
deserves further prospective association studies. It is
translationalmodificationssuchasglycosylationatly-
unlikelythatcancer-relatedweightlosscouldbeafac-
sine residues located in the collagenous domain of
tor in our patients since none of the patients reported
adiponectin are major determinants of the biological
significantweightloss. ThestudybyKaklamanietal.,
activityofadiponectin[41,42].
wasretrospectiveanddidnotincludedataontheBMI
In conclusion, several adiponectin polymorphisms
as well as other obesity-related metabolic parameters
have beenshown to influenceadiponectinlevels[12–
thatcouldinterferewiththegenotypeeffect[37].
14]. To date, few studies explored the association of
Discrepancies between our study and other associ-
adiponectin polymorphisms with obesity related can-
ation studies could also be explained by ethnic/racial
cers. Our study has shown that the genetic polymor-
differencesanditispossiblethatcertainallelesactually
phism SNP rs1501299 is an important genetic regu-
havedifferentfunctionsindifferentpopulations. Fur-
lator that results in increased adiponectin levels and,
thermore,differentSNPsinadiponectingenecangive
paradoxically,maybethepredisposingfactorinthese
rise to more than one allelic messenger ribose nucle-
patients. Furtherstudieswill be necessary to confirm
icacid(mRNA)formswhichinturnpossessdifferent
theroleofSNPrs1501299andSNPrs2241766inthe
biological functions as a result of differences in pri-
developmentofobesityrelatedcancers.
maryorhigherorderstructuresthatinteractwith oth-
ercellularcomponents[38]. Therecentfindingofin-
creasedlevelsoftissueadiponectininbreastcancerpa-
Acknowledgment
tientscomparedtocontrolsgivesfurthersupporttoour
findingof an association between an SNP thatcauses
higher adiponectin levels and cancer [39]. The study The financial supportreceived from the College of
by Karaduman et al., also reported that, women with Graduate Studies (Research Administration grant –
highlevelsoftissueadiponectinareatincreasedriskof YM11/07) and Kuwait Foundation for the Advance-
developingbreastcancer than womenwith low tissue ment of Science grant number (2006-1302-05)is ac-
adiponectinlevels [39]. Another recent study has re- knowledged.WeacknowledgecontributionoftheGen-
portedthatserumadiponectinlevelswerehigherinad- eralFacilityKUProject(GM01/05).
vancedthanorganconfinedprostatecancer[40]. This
study also reported that adiponectin could be used as
atumormarkertodifferentiatebetweenadvancedand
ConflictofInterest
localized stages of prostate cancer [40]. It is highly
probablethat patients with advancedstages of cancer
Noconflictofinterest.
couldhavehighercirculatingadiponectinespeciallyif
the cancer cells produce adiponectin. However there
arenostudiesthathaveevaluatedtissueexpressionof
adiponectininrelationtocirculatingadiponectinlevels Genesdiscussed
and no studies have evaluatedadiponectinproduction
fromtumorcells as a potentialtumormarker. Ifcon- Adiponectin-encodinggene: ADIPOQOrACDC
Description:female subjects having higher adiponectin levels than female subjects as shown in Table 2. Adiponectin- encoding gene: ADIPOQ Or ACDC